Published online Nov 6, 2020. doi: 10.12998/wjcc.v8.i21.5296
Peer-review started: July 13, 2020
First decision: August 8, 2020
Revised: September 8, 2020
Accepted: September 18, 2020
Article in press: September 18, 2020
Published online: November 6, 2020
We described the main features of an infant diagnosed with facial dysmorphic, language failure, intellectual disability and congenital malformations to strengthen our understanding of the disease. Currently, treatment is only rehabilitation and surgery for cleft lip and palate.
The proband was a 2-years-8-months-old girl. Familial history was negative for congenital malformations or intellectual disability. The patient had microcephaly, upward-slanting palpebral fissures, depressed nasal bridge, bulbous nose and bilateral cleft lip and palate. Brain magnetic resonance imaging showed cortical atrophy and band heterotopia. Her motor and intellectual development is delayed. A submicroscopic deletion in 11p13 involving the elongator acetyltransferase complex subunit 4 gene (ELP4) and a loss of heterozygosity in Xq25-q26.3 were detected.
There is no treatment for the ELP4 deletion caused by a submicroscopic 11p3 deletion. We describe a second case of deletion of the ELP4 gene without aniridia, which confirms the association between ELP4 gene with several defects and absence of this ocular defect. Additional clinical data in the deletion of the ELP4 gene as cleft palate, facial dysmorphism, and changes at level brain could be associated to this gene or be part of the effect of the recessives genes involved in the loss of heterozygosity region of Xq25-26.3.
Core Tip: We report a case diagnosed with submicroscopic 11p13 deletion. The main clinical characteristics and elongator acetyltransferase complex subunit 4 gene deletion, and treatments were assessed and a review of the related literature was performed. Very important, this is the second case of deletion of the elongator acetyltransferase complex subunit 4 gene without aniridia.
- Citation: Toral-Lopez J, González Huerta LM, Messina-Baas O, Cuevas-Covarrubias SA. Submicroscopic 11p13 deletion including the elongator acetyltransferase complex subunit 4 gene in a girl with language failure, intellectual disability and congenital malformations: A case report . World J Clin Cases 2020; 8(21): 5296-5303
- URL: https://www.wjgnet.com/2307-8960/full/v8/i21/5296.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i21.5296
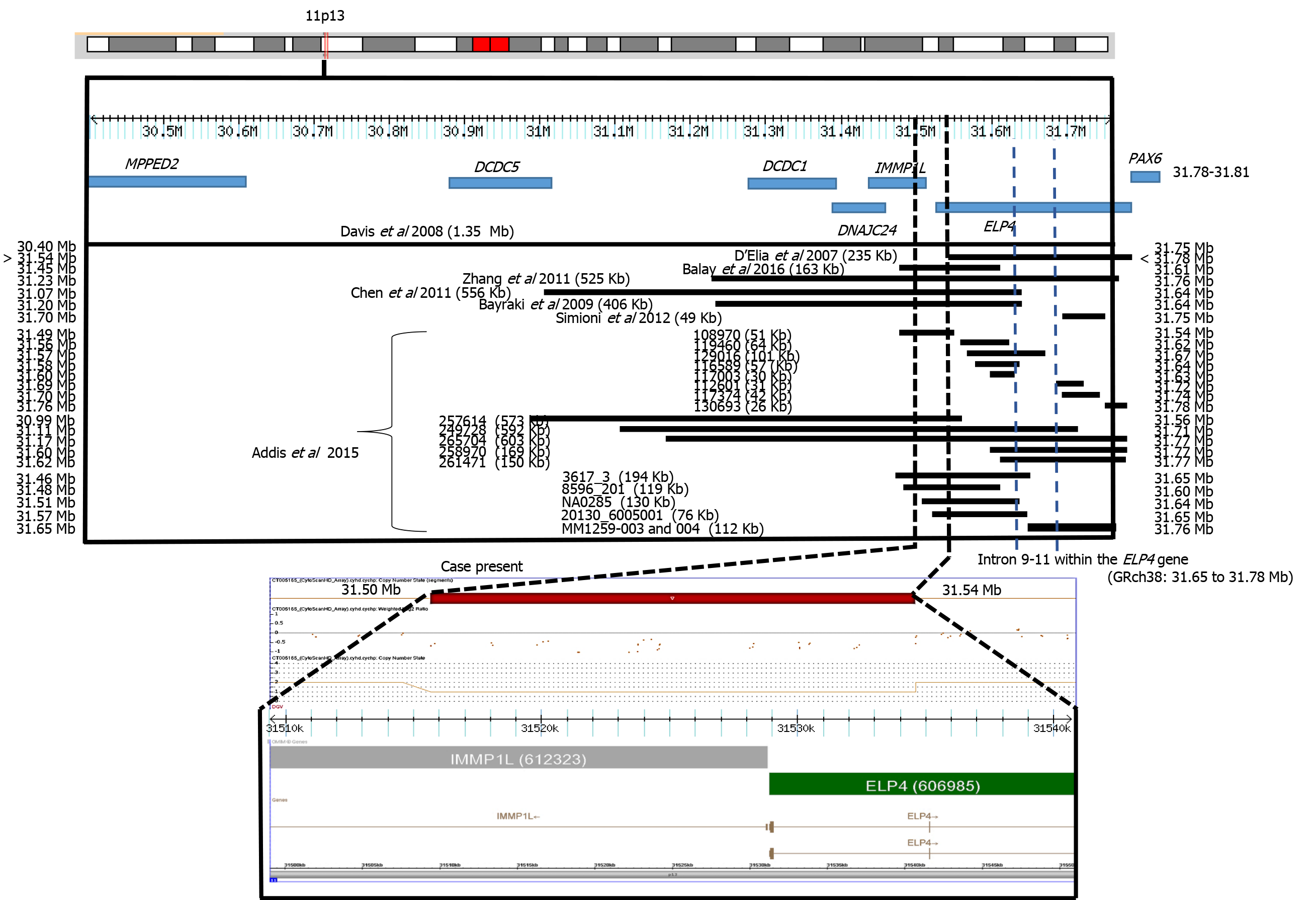
The elongator acetyltransferase complex subunit 4 gene (ELP4; MIM #606985) encodes the protein 4 of the elongator complex of ribonucleic acid polymerase II. ELP4 protein is composed of 424 amino acids and plays a role in transcriptional elongation, transfer ribonucleic acid modification, polarized exocytosis, and multiple types of cell migration. The failure of any member of the family of elongators, including ELP4, can be associated with different neurological disorders[1,2]. Twenty-five submicroscopic deletions, including the ELP4 gene and the adjacent sequences (excluding the PAX6 gene, the neighboring gene related to aniridia), have been associated with intellectual disability, language development failure, autism spectrum disorder, and epilepsy with aniridia[3-9] or without aniridia[10]. The ELP4 gene is 273.9 kb in size and encompasses 12 exons. Interestingly, inside the ultraconserved large intronic region between exons 9 to 12 of the ELP4 gene, there is a long-range cis-regulatory enhancer element located 25 to 150 kb downstream of the PAX6 gene, which controls its expression[11,12]. In the present study, we described a girl with dysmorphia, language failure, intellectual disability, and congenital malformations without aniridia, and with a submicroscopic deletion in 11p13 affecting the ELP4 gene.
Registering a cleft lip and palate at 26 wk of gestation and delayed motor development at 2 years of age.
The patient, a 2-year-and-8-month-old Mexican girl, was brought by her parents for evaluation because of delays in her motor and language development and congenital malformations. Currently, her motor development is abnormal without head control, she still does not sit down. She also does not speak any words and often becomes ill from the respiratory tract without any serious complications.
The proband was the third child of two healthy, unrelated, and young parents (27 and 26 years old at the time of delivery). Their familial history was negative for congenital malformations or intellectual disability. The mother had prenatal care, registering a cleft lip and palate at 26 wk of gestation. The proband was born by cesarean section at 38 wk of gestation with a weight of 3035 g (25th percentile), a length of 50 cm (25th–50th percentile), an OFC of 33 cm (10th percentile), and Apgar scores of 81 and 95. She did not require neonatal management.
Their familial history was negative for congenital malformations or intellectual disability.
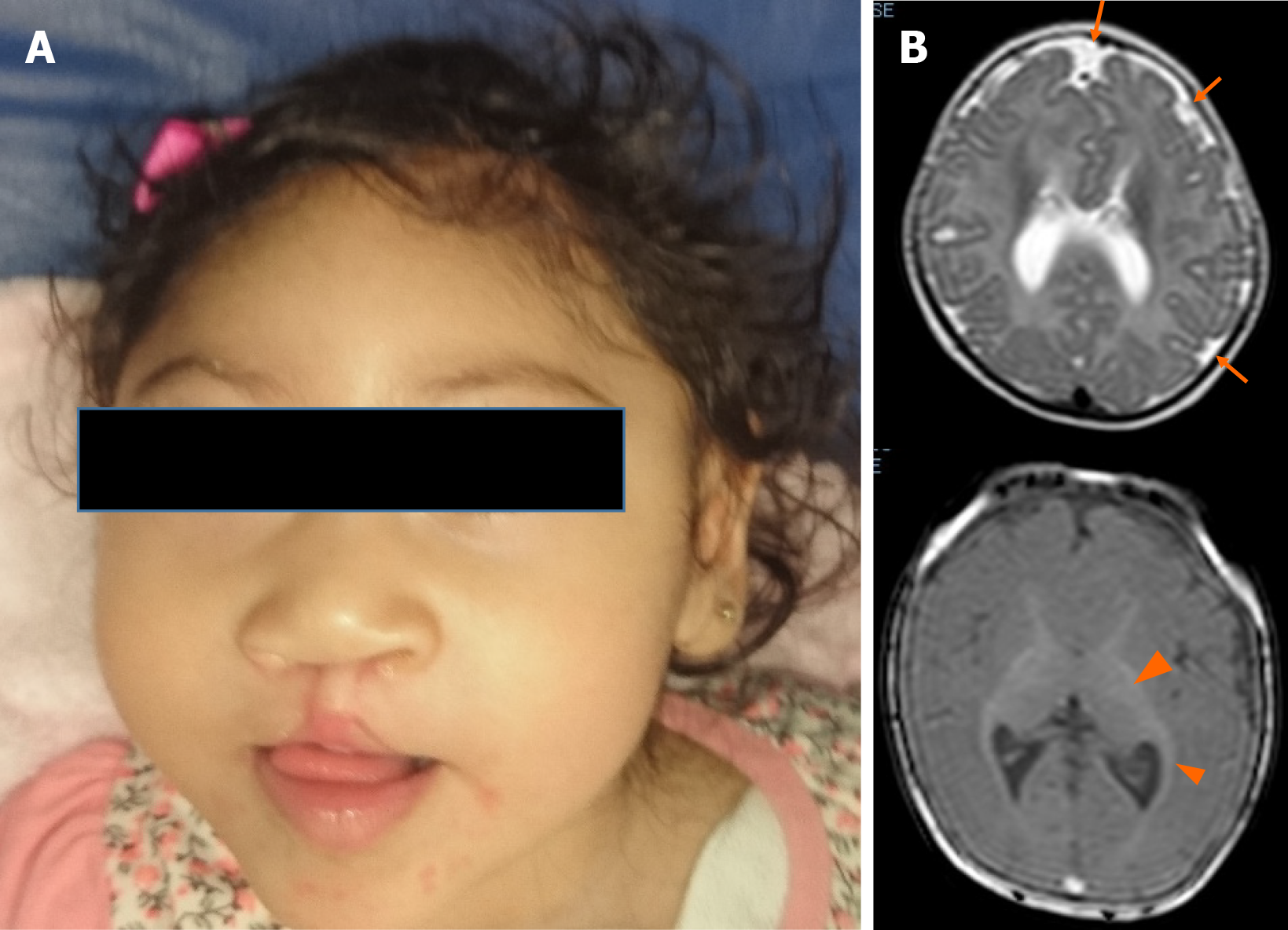
Upon physical examination, her weight was 9.2 kg (< 3rd percentile), her length was 87 cm (3rd–10th percentile), and her OFC was 46 cm (< 3rd percentile). She had microcephaly, upward-slanting palpebral fissures, a depressed nasal bridge, a bulbous nose, and a bilateral cleft lip and palate (Figure 1A).
Blood, urine, and thyroid profile analyses were normal. The karyotype was 46, XX.
The abdominal ultrasound was normal. The brain magnetic resonance imaging showed cortical atrophy, pachygyria, microgyria and band heterotopia (Figure 1B).
The auditory study detected neurosensory hearing loss. The ophthalmic and cardiological assessments were normal.
Genomic deoxyribonucleic acid from the proband and her parents was isolated from peripheral blood samples using the Gentra Pure Gene Blood Kit and Qiagen extraction kits. The oligonucleotide-single nucleotide polymorphism (SNP) array analysis with the GeneChip Human Cytoscan high definition was carried out for the patient and her parents following the provided protocol (Affymetrix, Santa Clara, Calif., United States) and using the Affymetrix GeneChip Scanner 3000 7G. The data were analyzed using GTYPE (GeneChip Genotyping Analysis Software, version 1.0.12) to detect copy number aberrations. The resolution of this procedure was estimated at 1.15 kb with 2.67 million probes. Copy number variation (CNV) breakpoints were determined by inspecting the log2 intensity ratios of SNPs within and flanking the detected regions of gain or loss.
The interpretation of the clinical significance of all the observed CNVs was compared with the database of genomic variants (https://projects.tcag.ca/variation/), the University of California Santa Cruz genome browser (http://genome.ucsc.edu/), Ensembl Resources, Online Mendelian Inheritance of human (OMIM), ClinGen, and ClinVar. The gene content of the CNVs of interest was determined with the University of California Santa Cruz University of California Santa Cruz browser based on the reference of the human genome national center of biotechnology information build 38 (hg38). For putative candidate regions containing at least one gene, the assessment included searches for similar cases in DECIPHER (https://decipher.sanger.ac.uk/) and a review in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/). The CNV pathogenicity was assessed using the described guidelines[13,14]. The interpretation depended on whether a given CNV overlapped with a known genomic disorder or was present in a patient with a similar phenotype or who was reported on the database of genomic variants database.
Submicroscopic 11p13 deletion involving the ELP4 gene with loss of heterozygosity in Xq25-26.3.
There is no specific treatment for the deletion of the ELP4 gene, the patient was managed with rehabilitation and surgery for cleft lip and palate.
No improvement or progress was observed as a result of the rehabilitation.
In many patients, the clinical presentation is not fully consistent with the syndrome under consideration and laboratory confirmation rates are low. In this study, we describe the use of molecular karyotype by SNP high definition arrays to investigate the presence of pathogenic CNV in a patient with several findings consistent with a syndrome of unknown etiology. A submicroscopic deletion of 31 kb was found to be present. This CNV was located in a region with recurrent submicroscopic deletions involving the ELP4 gene and subjacent regions (Figure 2). The ELP4 gene has been associated with intellectual disability, language development failure, autism spectrum disorder, epilepsy, and aniridia[3-9]. Our patient presented intellectual disability and language development failure but not autism spectrum disorder, epilepsy, or aniridia. These clinical data resemble those reported in a previous case with ELP4 gene deletion without aniridia. In this previous case, the family had a submicroscopic deletion of 163 kb in the ELP4 gene due to a pericentric inversion of chromosome 11p13. The patient presented intellectual disability, speech abnormalities, and autistic behaviors[10]. In another report, two families with bilateral aniridia, cataracts, and glaucoma had a deletion of 235 kb in the ELP4 gene[3]. Davis et al[4] also reported the case of a patient with aniridia, autism, and intellectual disability due to a 1354 kb deletion involving the ELP4 gene, whereas Bayrakli et al[5], studied a family with isolated aniridia that showed a deletion of 406 kb involving the ELP4 gene. A study in various members of a family with aniridia showed a 566 kb deletion involving the ELP4 gene[6], and Zhang et al[7] described a family with bilateral aniridia and congenital cataracts but without intellectual disability or other abnormalities due to a 525 kb deletion involving the ELP4 gene. Simioni et al[8] reported on a child with developmental delay, bilateral strabismus, aniridia, and nystagmus. The propositus showed a deletion of 49 kb in the ELP4 gene. Finally, Addis et al[9] made a comparison between 7235 cases and 11252 controls. The cases had language impairment, developmental delay, autism, and epilepsy. Thirteen cases presented submicroscopic deletions in 11p13. These researchers[9] reviewed DECIPHER and found 5 other cases with deletion, all overlapping with the ELP4 gene and suggesting a strong association between these deletions and neurodevelopmental disorders. All of these previous cases had deletions with a minimum distance of 10 kb and a maximum of 240 kb from the most proximal breakpoint to the 3′ end of the PAX6 gene, with an average of 103 kb (Figure 2).
Aniridia 2 is caused by mutations affecting a long-range cis-regulatory enhancer element of PAX6 expression inside of an ultra-conserved large intronic region between exons 9 to 12 of the ELP4 gene, located 25 to 150 kb downstream of the PAX6 gene[11]. These variants inside the ELP4 gene do not alter its normal expression and function[12]. The deletion in our patient did not include this region. Probably for this reason, the patient did not present aniridia, similar to the case of Balay et al[10]. Interestingly, the phenotypes of microcephaly, facial dysmorphia, cleft lip/ palate, neuromigration defect, and intellectual disability of our patient has been observed in the Baraitser-Winter cerebrofrontofacial syndrome 1 and 2 [OMIM 243310; BRWS1, 614583; BRWS2)][15], but the ACTB and ACTG1 genes of Baraitser-Winter cerebrofrontofacial syndrome 1 and Baraitser-Winter cerebrofrontofacial syndrome 2 were not involved in the deletion or duplication regions or loss of heterozygosity (LOH) of our patient. In a study of 117 cases with mental delay and/or congenital malformations, 434 CNVs (195 Losses and 239 gains), including 18 pathogenic and 9 potentially pathogenic, were found to be present. Interestingly, two patients with thrombocytopenia-absent-radius syndrome were not suspected by the clinicians, possibly because of the presence of atypical features. Two patients showed a pathogenic CNV with a syndrome that was neither manifesting nor suspected, demonstrating the difficulty in making accurate clinical diagnoses in some patients with classic microdeletion and microduplication syndromes and exemplifying an unexpected discovery of non-penetrant or presymptomatic conditions. In the aforementioned study, segmental regions of loss of heterozygosity larger than 5 Mb were found in 5 patients. An analysis of microsatellite markers within the segments of LOH was carried out in some cases, confirming homozygosity of biparental origin for these regions[16].
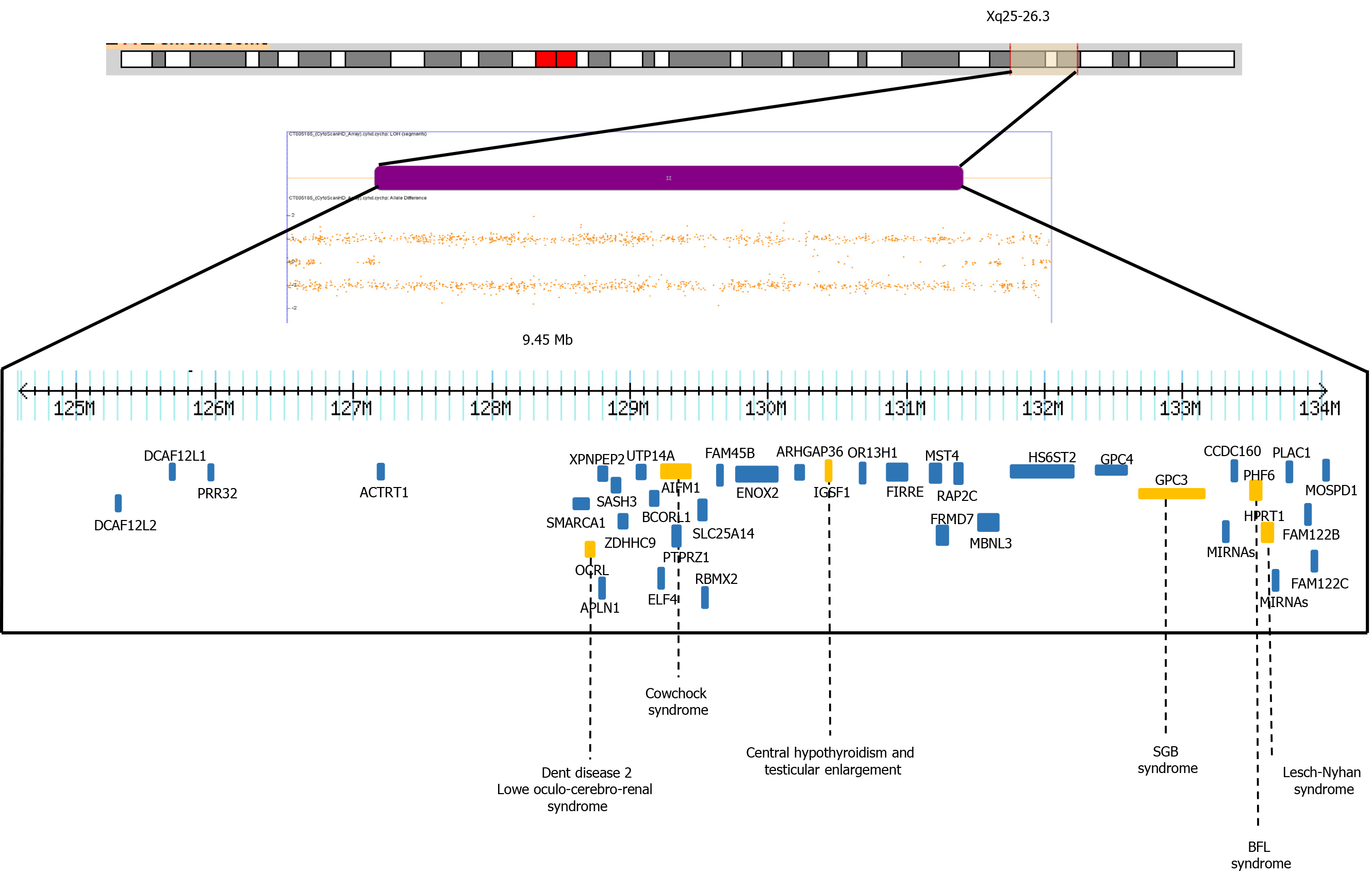
In our patient, the SNP microarray analysis also detected a segment of LOH of 9.4 Mb, which included 39 OMIM genes, four of them have been associated with intellectual disability, OCRL, AIFM1, PHF6 and HPRT1 genes (HP: 0001249 in OMIM) (Figure 3). The Borjeson-Forssman-Lehman syndrome syndrome has been associated with cleft lip/palate, microcephaly, band heteropia and simplified gyral cortical patterns as the case of some female due to a de novo intragenic duplication of
Any pathogenic recessive mutation within these regions could theoretically be clinically significant, and the likelihood of this occurring would increase with the number of regions present. However, as normal genome variations, it is unlikely that most of these regions are clinically significant. A previous study with 8 families identified long segments (from 2.2% to 4.4% of the autosomal genome) and short homozygosity using short tandem repeat markers, indicating that the parents were relatively related[19]. Another study with 209 unrelated individuals from a HapMap population detected 1393 segments over 1 Mb in length (the longest length was a region of 17.9 Mb). In the aforementioned study, the subjects had an average of 6.6 homozygous segments with larger long tracts in regions of linkage disequilibrium and low recombination. This suggests that multiple ancestral megabase haplotypes persist in non-inbred human populations in broad genomic regions with below-average recombination rates[20]. Finally, an analysis of 276 neurologically normal elderly subjects among North American Caucasians using the SNP of the entire genome revealed contiguous tracts of > 5 Mb in 9.5% (26/272) of these individuals, indicating that the long homozygous segments represent segments of autozygosity due to the mating of closely related individuals, while short segments were due to linkage disequilibrium or past ancestors who were subject to distant inbreeding[21].
Microsatellite analysis in our patient’s parents could confirm the homozygosity of the biparental origin for these regions. It is important to mention that in women the X inactivation mechanism compensates for these types of molecular changes, minimizing or nullifying the phenotypic effects. To interpret LOH, it is necessary to add a catalog of new pathogenic or potentially pathogenic loci in high-quality database. This could provide the opportunity to perform genotype–phenotype correlations in a suitable number of individuals with congenital malformations and/or intellectual disability.
In conclusion, we describe a second case of deletion of the ELP4 gene, which confirms its association with the absence of aniridia and the presence of neurological manifestations. We also showed additional clinical data on cleft palate/Lip or facial dysmorphism and changes at the brain level, probably due to the affectation of the genes involved in the LOH region. More case reports or large studies involving patients with the ELP4 gene are necessary.
The authors thank the family for participating in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vieira A, Wattanasirichaigoon D S-Editor: Zhang L L-Editor: A P-Editor: Wang LL
| 1. | Nguyen L, Humbert S, Saudou F, Chariot A. Elongator - an emerging role in neurological disorders. Trends Mol Med. 2010;16:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 2. | Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, Neale BM, Turner MR, Veldink JH, Ophoff RA, Tripathi VB, Beleza A, Shah MN, Proitsi P, Van Hoecke A, Carmeliet P, Horvitz HR, Leigh PN, Shaw CE, van den Berg LH, Sham PC, Powell JF, Verstreken P, Brown RH Jr, Robberecht W, Al-Chalabi A. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 203] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 3. | D'Elia AV, Pellizzari L, Fabbro D, Pianta A, Divizia MT, Rinaldi R, Grammatico B, Grammatico P, Arduino C, Damante G. A deletion 3' to the PAX6 gene in familial aniridia cases. Mol Vis. 2007;13:1245-1250. [PubMed] [Cited in This Article: ] |
| 4. | Davis LK, Meyer KJ, Rudd DS, Librant AL, Epping EA, Sheffield VC, Wassink TH. Pax6 3' deletion results in aniridia, autism and mental retardation. Hum Genet. 2008;123:371-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Bayrakli F, Guney I, Bayri Y, Ercan-Sencicek AG, Ceyhan D, Cankaya T, Mason C, Bilguvar K, Bayrakli S, Mane SM, State MW, Gunel M. A novel heterozygous deletion within the 3' region of the PAX6 gene causing isolated aniridia in a large family group. J Clin Neurosci. 2009;16:1610-1614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Cheng F, Song W, Kang Y, Yu S, Yuan H. A 556 kb deletion in the downstream region of the PAX6 gene causes familial aniridia and other eye anomalies in a Chinese family. Mol Vis. 2011;17:448-455. [PubMed] [Cited in This Article: ] |
| 7. | Zhang X, Zhang Q, Tong Y, Dai H, Zhao X, Bai F, Xu L, Li Y. Large novel deletions detected in Chinese families with aniridia: correlation between genotype and phenotype. Mol Vis. 2011;17:548-557. [PubMed] [Cited in This Article: ] |
| 8. | Simioni M, Vieira TP, Sgardioli IC, Freitas EL, Rosenberg C, Maurer-Morelli CV, Lopes-Cendes I, Fett-Conte AC, Gil-da-Silva-Lopes VL. Insertional translocation of 15q25-q26 into 11p13 and duplication at 8p23.1 characterized by high resolution arrays in a boy with congenital malformations and aniridia. Am J Med Genet A. 2012;158A:2905-2910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Addis L, Ahn JW, Dobson R, Dixit A, Ogilvie CM, Pinto D, Vaags AK, Coon H, Chaste P, Wilson S, Parr JR, Andrieux J, Lenne B, Tumer Z, Leuzzi V, Aubell K, Koillinen H, Curran S, Marshall CR, Scherer SW, Strug LJ, Collier DA, Pal DK. Microdeletions of ELP4 Are Associated with Language Impairment, Autism Spectrum Disorder, and Mental Retardation. Hum Mutat. 2015;36:842-850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Balay L, Totten E, Okada L, Zell S, Ticho B, Israel J, Kogan J. A familial pericentric inversion of chromosome 11 associated with a microdeletion of 163 kb and microduplication of 288 kb at 11p13 and 11q22.3 without aniridia or eye anomalies. Am J Med Genet A. 2016;170A:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | McBride DJ, Buckle A, van Heyningen V, Kleinjan DA. DNaseI hypersensitivity and ultraconservation reveal novel, interdependent long-range enhancers at the complex Pax6 cis-regulatory region. PLoS One. 2011;6:e28616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Bhatia S, Bengani H, Fish M, Brown A, Divizia MT, de Marco R, Damante G, Grainger R, van Heyningen V, Kleinjan DA. Disruption of autoregulatory feedback by a mutation in a remote, ultraconserved PAX6 enhancer causes aniridia. Am J Hum Genet. 2013;93:1126-1134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 13. | Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 600] [Cited by in F6Publishing: 648] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 14. | South ST, Lee C, Lamb AN, Higgins AW, Kearney HM; Working Group for the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med. 2013;15:901-909. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 222] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 15. | Verloes A, Di Donato N, Masliah-Planchon J, Jongmans M, Abdul-Raman OA, Albrecht B, Allanson J, Brunner H, Bertola D, Chassaing N, David A, Devriendt K, Eftekhari P, Drouin-Garraud V, Faravelli F, Faivre L, Giuliano F, Guion Almeida L, Juncos J, Kempers M, Eker HK, Lacombe D, Lin A, Mancini G, Melis D, Lourenço CM, Siu VM, Morin G, Nezarati M, Nowaczyk MJ, Ramer JC, Osimani S, Philip N, Pierpont ME, Procaccio V, Roseli ZS, Rossi M, Rusu C, Sznajer Y, Templin L, Uliana V, Klaus M, Van Bon B, Van Ravenswaaij C, Wainer B, Fry AE, Rump A, Hoischen A, Drunat S, Rivière JB, Dobyns WB, Pilz DT. Baraitser-Winter cerebrofrontofacial syndrome: delineation of the spectrum in 42 cases. Eur J Hum Genet. 2015;23:292-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 16. | Bruno DL, Ganesamoorthy D, Schoumans J, Bankier A, Coman D, Delatycki M, Gardner RJ, Hunter M, James PA, Kannu P, McGillivray G, Pachter N, Peters H, Rieubland C, Savarirayan R, Scheffer IE, Sheffield L, Tan T, White SM, Yeung A, Bowman Z, Ngo C, Choy KW, Cacheux V, Wong L, Amor DJ, Slater HR. Detection of cryptic pathogenic copy number variations and constitutional loss of heterozygosity using high resolution SNP microarray analysis in 117 patients referred for cytogenetic analysis and impact on clinical practice. J Med Genet. 2009;46:123-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Gécz J, Turner G, Nelson J, Partington M. The Börjeson-Forssman-Lehman syndrome (BFLS, MIM #301900). Eur J Hum Genet. 2006;14:1233-1237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Kasper BS, Dörfler A, Di Donato N, Kasper EM, Wieczorek D, Hoyer J, Zweier C. Central nervous system anomalies in two females with Borjeson-Forssman-Lehmann syndrome. Epilepsy Behav. 2017;69:104-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain. Am J Hum Genet. 1999;65:1493-1500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 200] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 20. | Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789-795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 278] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 21. | Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 184] [Article Influence: 10.2] [Reference Citation Analysis (0)] |