Abstract
Maternal pregnancy levels of the inflammatory marker C-reactive protein (CRP) has been previously associated with autism spectrum disorder (ASD) in the offspring. We conducted a population-based nested case–control study with 500 children with ASD, 235 with developmental delay (DD) and 580 general population (GP) controls to further investigate whether elevated CRP during pregnancy increases the risk of ASD. Maternal CRP concentration was measured in archived serum collected during 15–19 weeks of pregnancy and genome-wide single-nucleotide polymorphism (SNP) data were generated. The levels of CRP were compared between ASD vs GP and DD vs GP. The genetic associations with CRP were assessed via linear regression. Maternal CRP levels in mid-pregnancy were lower in mothers of ASD compared with controls. The maternal CRP levels in the upper third and fourth quartiles were associated with a 45 and 44% decreased risk of ASD, respectively. Two SNPs at the CRP locus showed strong association with CRP levels but they were not associated with ASD. No difference was found between maternal CRP levels of DD and controls. The reasons for the lower levels of CRP in mothers of ASD are not known with certainty but may be related to alterations in the immune response to infectious agents. The biological mechanisms underlying this association remain to be clarified.
Similar content being viewed by others
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with unknown cause(s) for the vast majority of cases. Previous studies suggest that both genetic1, 2, 3, 4 and environmental5, 6 factors contribute to the etiology of autism. However, the amount that each factor contributes is the subject of an ongoing debate.7, 8, 9 Among the suspected environmental factors of autism, maternal infections during pregnancy have been reported by a number of epidemiological studies.10, 11, 12, 13, 14, 15 The studies using animal models have also shown an association between maternal infection during pregnancy and abnormal behaviors in offspring16, 17 and suggested that maternal immune activation might be one pathway by which maternal infection can lead to elevated risk of autism in the offspring.18 Fever, an acute inflammatory response to various environmental factors including infections, has been suggested to increase the risk of autism and developmental delays.10, 14
Acute inflammation is a multisystem process that occurs following injury and/or infection. During this process, immune cells release cytokines, including interleukin (IL)-1, IL-6, tumor necrosis factor-alpha, interferon gamma and others. Such pro-inflammatory cytokines in turn induce the liver to synthesize various acute-phase reactant proteins including C-reactive protein (CRP). The levels of CRP increase very rapidly in response to trauma, inflammation and infection and decrease just as rapidly with the resolution of the condition.19 Thus, the measurement of CRP is widely used to monitor inflammatory states, including those that have been suggested to increase the risk of autism.20
In addition to variation with inflammatory states, the variation in human levels of CRP has been estimated to be 35–40% heritable in several twin studies.21 To identify the genetic determinants of CRP levels, genome-wide association study (GWAS) approaches have been used: the strongest influence on CRP levels is the locus containing the CRP gene itself. However, additional loci implicate metabolic pathways as well as innate and adaptive immune responses, including associations near the IL6, IL6R, NLRP3, IL1F10 and IRF1 genes.22, 23, 24
Because association does not always indicate causation, previous studies have utilized a Mendelian randomization approach to leverage genetic information to assess the possible causal relationship of CRP levels with health outcomes. This approach takes advantage of the random allocation of alleles at conception to test the hypothesis that a trait is causally linked to disease. To support causality, the genetic variants that influence the trait should also influence disease risk. For example, this approach has shown that CRP concentration is associated but not causal for coronary heart disease.25, 26
Increased levels of CRP during pregnancy have been associated with a number of adverse pregnancy outcomes including preterm birth, low birth weight, small for gestational age and pre-eclampsia.27, 28, 29, 30, 31 However, the association between CRP levels during pregnancy and autism has not been intensively studied. The only published study, to our knowledge, reported that elevated levels of CRP (>5.84 mg dl−1) during the first and early second trimesters of pregnancy increased risk of ASD by 43%.32 However, the study did not investigate whether maternal CRP levels were also associated with other developmental disorders.
The objective of this study was to further examine whether maternal mid-pregnancy CRP levels are associated with increased risk of autism and other developmental delays and whether this association might be influenced by genetic determinants of CRP levels.
Materials and methods
Study population
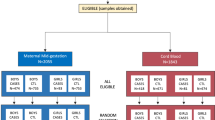
The samples for the present study were derived from the early markers for autism study, a population-based nested case–control study designed to evaluate the biologic markers of susceptibility and exposure in archived maternal mid-pregnancy and neonatal blood specimens from the same mother–baby pairs. The early markers for autism study population was drawn from the cohort of children born in California from 2000 to 2003 to women who were pregnant in Orange, San Diego and Imperial Counties, CA, USA, and who participated in the State’s prenatal expanded alpha-fetoprotein screening program. Among women delivering live births in the study counties and birth years, 70% had prenatal expanded alpha-fetoprotein screening program screening. The characteristics of these screened women were similar to the characteristics of all the women in the study counties and birth years with respect to maternal age, education level, race/ethnicity, place of birth, parity and health insurance status (Supplementary Appendix Table 1). Three groups of children were identified: children with ASD, children with developmental delay (DD) but not ASD, and general population controls (GP). Children with ASD or DD were ascertained from the California Department of Developmental Services, which operates a system of 21 regional centers that coordinate services for persons with ASD, mental disability and other developmental disabilities. The GP controls were randomly sampled from the birth certificate files after excluding all past or current department of developmental service/regional center clients and were frequency matched to ASD cases by sex, birth month and birth year.
The early markers for autism study samples were obtained in two phases resulting in two independent sample sets. The first phase included 84 children with ASD, 47 DD and 148 GP children born in 2000 and 2001 in the Orange County. The second phase included 416 ASD, 188 DD and 432 GP children born between 2000 and 2003 in the Orange, San Diego and Imperial Counties, CA, USA. The children in the first phase are not included in the second phase.
Diagnostic verification
ASD and DD diagnoses were verified by a developmental pediatrician following the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition criteria by review of information abstracted from medical records following a protocol initially developed by the Metropolitan Atlanta Developmental Disabilities Surveillance Program.33 Diagnostic agreements between the developmental pediatrician and the department of developmental services were >90% for ASD and >50% for DD.
CRP measurements
The maternal mid-pregnancy (15–19 weeks gestation) blood specimens were retrieved from the leftover samples of the prenatal screening specimen archives maintained by the Genetic Disease Screening Program, California Department of Public Health. The CRP measurements for each phase of early markers for autism study samples were conducted at a different laboratory using a different technique. The samples for the first phase were analyzed at Dr Yolken’s laboratory at John Hopkins University using a microplate solid phase immunoassay method. The reagents were obtained from IBL America, Minneapolis, MN, USA and the assays were performed as previously described.34, 35 Duplicate standards were run on each assay plate and the values below the detection limit were analyzed as 0. The sensitivity of the assay allowed the detection of CRP levels greater than 500 ng ml−1. The samples for the second phase were analyzed at Dr Van de Water laboratory at the University of California, Davis via a Milliplex magnetic bead immunoassay (Millipore, Billerica, MA, USA) that utilized the Luminex fluorescent-bead-based technology. The CRP concentrations were calculated with the Bio-Plex Manager software using standard curves derived from the known reference cytokine concentrations supplied by the manufacturer. The sensitivity of this assay allowed the detection of CRP at levels greater than 400ng ml−1 and values below the limit of detection were assigned the value of (limit of detection)/2. The Luminex method has been used in numerous previous studies suggesting that the method is both reliable and reproducible.20, 32, 36 All the laboratory assays were conducted blinded to case–control status.
DNA extraction and genotyping
The maternal ASD and GP blood samples from the Phase 2 set were treated for DNA extraction as described in Tsang et al.37 All the maternal samples were genotyped using the Affymetrix Axiom EUR array by the Genomics Core Facility (GCF) at UCSF, using standard protocols. The Axiom EUR array assays approximately 675 000 single-nucleotide polymorphisms (SNPs) across the genome, and is optimized for genome-wide, gene-based and candidate-SNP coverage.38 The genotype calling was carried out using Affymetrix Power-Tools in accordance with the Axiom Genotyping Solution Data Analysis Guide provided by Affymetrix (http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx.). The quality controls were carried out for the genotypes and samples to be included in analysis using PLINK software39 as reported in Tsang et al.37
Statistical analysis
Association between maternal CRP and ASD
We analyzed the two samples separately because they were processed in two different laboratories using two different techniques. We compared sociodemographic factors between ASD, DD and GP groups using chi-square tests for categorical variables and t-tests for continuous variables. Among the GP group, we also determined the association between sociodemographic factors and maternal CRP levels. The sociodemographic factors were: child sex, child birth month (calendar month), birth year; and maternal ethnicity, race, place of birth (USA, Mexico, other), age at child birth, weight at blood draw and level of education.
The maternal CRP was first analyzed as a continuous variable after natural log-transformation to normalize the distribution, and then categorized into quartiles and quintiles based on the distribution in the GP group. We conducted crude and adjusted logistic regression analyses to determine the association between maternal CRP levels and ASD or DD. In all categorical analyses, the lowest quartile and quintile were the reference group. Sociodemographic factors associated with both ASD and maternal CRP at P=0.05 were used to adjust the crude odds ratio in addition to the matching variables (child sex, birth month and birth year). We also tested whether adjustment for ancestry, as measured by principal components (PCs) derived from GWAS data, altered the results. PC-adjusted models included other potential confounders described above, with the exception of race/ethnicity, which would be accounted for by the PCs. To further explore the association between CRP and ASD, we examined the risk of ASD in the children of mothers with CRP levels ⩾1 mg dl−1, a concentration associated with clinical relevance.19
CRP heritability and genome-wide association analysis
The final set of high-quality SNP markers from ASD and GP control mothers from the phase 2 sample set was used to generate a genetic-relationship matrix and calculate marker-based additive heritability (h2) of log-transformed CRP levels after exclusion of outliers. A restricted maximum-likelihood model implemented in GCTA software40 was utilized with the same sociodemographic covariates as above, plus 10 genetic ancestry principal components and a covariate indicating child phenotype (ASD, DD, GP). The heritability estimate indicates the proportion of the total phenotypic variance in CRP level explained by the polygenic genetic contribution estimated by genotyped markers after the exclusion of the effect of sociodemographic factors and of 10 genetic ancestry principal components.
A GWAS study was performed on CRP levels for the phase 2 ASD and GP control mother set with a linear regression analysis for common SNPs adjusted for the same set of covariates using PLINK software.39 The distribution of test statistics has been evaluated with a quantile–quantile plot generated in R 3.2.0 environment (The R Project for Statistical Computing [http://www.r-project.org]) and the top associated genetic loci with a marker that reached genome-wide significance after Bonferroni multiple test correction (cutoff P=7.6 × 10−8), were considered. Linear and logistic regression models were used to assess the significance of the GWAS top SNPs in a model for CRP levels and for ASD outcome in the R 3.2.0 environment. The linkage disequilibrium among the genetic markers located in the genomic region of associated top SNPs was assessed measuring r2 using the 1000 Genome Project EUR genetic data as the reference panel.41 The regional genomic plots that visualize in a log-scale the association P-value and the reciprocal r2 between the top hit and other genetic markers were drawn using Locuszoom tool.42
The study was approved by the institutional review boards of the California Health and Human Services Agency and Kaiser Permanente of Northern California.
Results
Sociodemographic factors
The first sample included 84 children with ASD, 46 with DD and 148 GP children. There was no difference between the ASD cases and the GP controls for gestational age and maternal weight at blood draw. However, the mothers of ASD cases were more likely to be non-Hispanic, born in the United States and to be slightly older compared with the mothers of GP controls (Table 1). The second sample included 416 ASD cases, 188 DD and 432 GP. There was no difference between the ASD cases and the GP controls in maternal age, race, gestational age and maternal weight at blood draw. In sample set #1 and #2, maternal ethnicity, race, weight at blood draw and place of birth were associated with level of CRP (Supplementary Appendix Table 2). The mothers of ASD cases were slightly more educated, more likely to be non-Hispanic and born in the United States (Table 1). Only maternal ethnicity was associated with both ASD and CRP at P<0.05 but in our final model estimating risk of ASD associated with maternal CRP, we included maternal ethnicity, the matching variables (child sex, birth month and birth year) and maternal weight at blood draw because of its high association with CRP levels.
Association between maternal CRP and developmental outcome
First sample set
Median CRP levels were lower in the mothers of children with ASD (1.28 mg dl−1, interquartile range=0.54–3.06) compared with the mothers of GP controls (2.43 mg dl−1, interquartile range=0.61–3.82; Supplementary Appendix Figure 1A). We did not find an association between the maternal CRP levels (measured on a continuous scale) and the risk of ASD (crude odds ratio (OR)=0.88, 95% confidence interval (CI)=0.73–1.05; adjusted odds ratio (AOR)=0.97, 95% CI=0.78–1.20). There was no significant difference in the levels of maternal CRP between ASD and GP controls when comparing quartiles or quintiles. We found a similar null result comparing the DD with the GP controls (Table 2).
Second sample set
Median CRP levels were significantly lower in the mothers of children with ASD (1.94 mg dl−1, interquartile range=1.04–3.90) compared with the mothers of GP controls (2.40 mg dl−1, interquartile range=1.32–4.48; Supplementary Appendix Figure 1B). The risk of ASD decreased by 17% for every one unit increase in the level of maternal CRP during pregnancy (OR=0.83, 95% CI=0.73–0.95; AOR=0.83, 95% CI=0.72–0.97; Table 3). The proportion of women with a CRP concentration in the upper third and fourth quartiles was lower among the mothers of children with ASD compared with the mothers of GP controls (third quartile: 19.7% vs 25.3, AOR=0.57, 95% CI=0.38–0.85; fourth quartile: 20.9% vs 27.1%, AOR=0.58, 95% CI=0.38–0.89). We found a similar trend when CRP concentration was categorized into quintiles (Table 3). These results were adjusted for sex, birth month, birth year, maternal ethnicity and weight at blood draw. They remain unchanged if we replace maternal ethnicity with the derived 10 principal components representing genetic ancestry. The CRP values above 1 mg dl−1 were not associated with increased risk of ASD (data not shown). There was no significant difference in the levels of maternal CRP between DD and GP controls analyzing CRP as a continuous or categorical variable (Table 3).
To determine whether genetic determinants of CRP levels might be driving the association between CRP and ASD, we first assessed the overall genetic signal from the GWAS by examining whether the distribution of test statistics deviated from that expected by chance. In a quantile–quantile plot comparing observed with expected distributions, there was an elevated tail indicating strong genetic signal (Figure 1). The SNP association, which remained significant after Bonferroni multiple testing correction, was within the CRP locus itself, along with additional SNPs with suggestive association (P<10−6). The linkage disequilibrium in the region suggests two semi-independent signals (regional Supplementary Figure S1) (rs3116656; beta=0.30; P-value=1.4 × 10−8 and rs2794520; beta=−0.25; P-value=3.9 × 10−7; linkage disequilibrium r2<0.2). The two SNPs were associated with CRP levels regardless of case–control status (Table 4). After excluding the CRP locus, the distribution of test statistics returned to the null expectation (Figure 1) and there was no detectable polygenic SNP heritability for CRP levels (h2=0; s.e.=0.31). Thus, genetic determination of CRP levels can be adequately summarized by using these two SNP markers.
Quantile–quantile plots of the association of genome-wide single-nucleotide polymorphisms (SNPs) with maternal mid-pregnancy C-reactive protein (CRP) levels: SNPs located in CRP locus have been included (filled circles) or excluded (open triangles). The y axis shows the negative logarithm of the observed association P-value for each tested SNP; the x axis shows the negative logarithm of the expected P-value based on a statistical sample. The red line is the line of equivalence, observed = expected. The blue line shows the Bonferroni significance threshold for multiple tests (0.05/658 581 SNPs=7.6 × 10−8).
Incorporating the two SNPs representing the top loci into the regression model attenuated the association between the CRP level and the ASD outcome, but the association remained significant and negative (AOR=0.86, 95% CI=0.77–0.97). When we included an interaction term between CRP and SNPs in the regression models estimating risk of ASD, the P-values for the interaction terms were all greater than 0.1 (P-value for SNP1 × CRP=0.71 and P-value for SNP2 × CRP=0.22). The SNPs that significantly influence the CRP level were not associated with ASD (rs3116656: AOR=0.98, 95% CI=0.92–1.04; rs2794520: AOR=1.02, 95% CI=0.96–1.08).
Discussion
In the present study, we found no association between maternal mid-pregnancy CRP levels and risk of ASD in the smaller sample, but in the larger sample, the mothers of children with ASD had lower levels of mid-pregnancy CRP compared with those of GP controls. This finding is in contrast to the one prior study that examined CRP levels in association with ASD, which reported higher levels of CRP in the mothers of children with ASD compared with controls.32 Our current study and the study by Brown et al.32 have conflicting conclusions, even though the ranges of CRP levels were similar; the samples were collected at approximately the same period during pregnancy and both the studies used a similar laboratory technique to analyze the serum samples (our sample set 2). In contrast to the Brown study, which was based on an ethnically homogeneous sample from Finland, our sample included a racially and ethnically diverse population. Although the CRP levels have been shown to vary significantly across different ethnic groups,23 our finding of a negative association between the maternal CRP level and ASD was observed consistently within each race/ethnic group in our sample. Also in contrast to the Brown study, our study controlled for maternal weight at the time of blood draw, which is also associated with the CRP levels. However, maternal weight did not influence the direction of association that we observed.
Although there is no established consistent baseline levels of CRP, the levels have been shown to increase during normal pregnancy,29, 43, 44, 45 and the relationship between CRP level and ASD risk may also vary over the pregnancy. Our results reflect levels of CRP measured between gestational weeks 15–19 weeks only; we were unable to evaluate different pregnancy time periods. One study reported that values above 1 mg dl−1 during pregnancy are clinically abnormal.45 In our study, we did not find an association between those levels and increased risk of ASD. Hwang et al.29 reported that CRP levels ranged from 0.04 to 2.03 mg dl−1 during the 15–20 weeks of pregnancy, corresponding to the period during pregnancy when maternal samples were collected for our study. Among 99% of control group mothers from the second sample, the CRP values ranged from 0.04 to 22.2 mg dl−1. The 10-fold difference in the range of CRP levels between the two studies may be related to the differences in the assays used or other factors related to the study populations.
Further, we investigated whether genetic determinants of CRP levels might explain our association results or establish causality via Mendelian randomization. We found two significant associations with maternal CRP level at the CRP gene locus: the best hit rs3116656 is in high linkage disequilibrium (r2>0.8) with a previously reported CRP-associated SNP in Ridker et al.24 and the second, rs2794520, replicated a previous finding of Ridker et al. These semi-independent SNPs accounted for all the genetic signals that we observed, but did not account for the association between CRP and ASD outcome. Further, the CRP-associated SNPs were not themselves associated with ASD outcome. Thus, an unmeasured factor is likely to be associated with the CRP levels and driving the negative association that we observed between CRP and ASD. However, our Mendelian randomization analysis did not have sufficient power to determine causality for the observed association between CRP and ASD.
In the present study, we did not find an association between maternal mid-pregnancy CRP levels and developmental delay. These findings may suggest that variation in CRP levels during pregnancy may lead to changes that are specific to ASD. However, more studies are needed to confirm the specificity of the association between maternal levels of CRP and ASD.
Infections are the leading cause of acute inflammation. Previously, we found that maternal infections diagnosed in a hospital setting, which may represent cases of more severe infections, were associated with increased risk of ASD compared with infections diagnosed in an outpatient setting.15 Atladotir et al.10 also found that maternal fever lasting more than 7 days was associated with increased risk of ASD as compared with fever that lasted less than 7 days, suggesting that longer periods of inflammation are associated with increased risk of autism. The measured levels of CRP in the present study are more likely to represent baseline CRP at the time of blood collection, as they were normally distributed. The low levels of CRP in the mothers of children with ASD might indicate a possible immune dysregulation. Our present results combined with the previous results of association between infections and autism suggest that some mothers of children with ASD either mount an excessive inflammatory response to environmental factors or they have a baseline immune function that is low and do not mount the appropriate inflammatory response to clear infections or other environmental insults. However, more studies are needed to support these hypotheses.
The results of this study should be considered in light of the following limitations. We were unable to validate the diagnostic status of ASD by systematic clinical evaluation. Instead, we relied on expert review of information recorded as part of diagnostic eligibility for developmental services from regional center records. In addition, we did not have any clinical information such as infection status of the mothers and we were unable to stratify our analysis on maternal obesity status because the variable was not available. Another limitation of the study is that we were not able to stratify our analyses by ASD subtypes. The strengths of the study include the replication of results (although power was not sufficient in the first sample set, the trend was in the same direction as the significant findings in the second sample set) in two separate study populations with samples analyzed in two different laboratories using two distinct methods, and the investigation of genetic determinants of CRP level. All the laboratory assays were conducted blinded to case–control status. Finally, the inclusion of the DD group allowed us to evaluate the specificity of the findings to ASD.
Conclusion
The maternal CRP levels at 15–19 weeks gestation were lower in the mothers of ASD compared with controls. Although strong genetic determinants of CRP level exist, they do not account for the association between CRP and ASD outcome. The reasons for the lower levels of CRP in the mothers of ASD are not known with certainty but may be related to alterations in the immune response to infectious agents. The mechanisms behind these alterations should be the subject of future investigations.
References
Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995; 25: 63–77.
Folstein S, Rutter M . Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 1977; 18: 297–321.
Geschwind DH . Autism: many genes, common pathways? Cell 2008; 135: 391–395.
Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 1989; 30: 405–416.
Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN . The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect 2006; 114: 1119–1125.
Volk HE, Kerin T, Lurmann F, Hertz-Picciotto I, McConnell R, Campbell DB . Autism spectrum disorder: interaction of air pollution with the MET receptor tyrosine kinase gene. Epidemiology 2014; 25: 44–47.
Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry 2015; 72: 415–423.
Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB et al. Most genetic risk for autism resides with common variation. Nat Genet 2014; 46: 881–885.
Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry 2011; 68: 1095–1102.
Atladottir HO, Henriksen TB, Schendel DE, Parner ET . Autism after infection, febrile episodes, and antibiotic use during pregnancy: an exploratory study. Pediatrics 2012; 130: e1447–e1454.
Atladottir HO, Thorsen P, Ostergaard L, Schendel DE, Lemcke S, Abdallah M et al. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J Autism Dev Disord 2010; 40: 1423–1430.
Chess S . Autism in children with congenital rubella. J Autism Child Schizophr 1971; 1: 33–47.
Lee BK, Magnusson C, Gardner RM, Blomstrom S, Newschaffer CJ, Burstyn I et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav Immun 2014; 44: 100–105.
Zerbo O, Iosif AM, Walker C, Ozonoff S, Hansen RL, Hertz-Picciotto I . Is maternal influenza or fever during pregnancy associated with autism or developmental delays? Results from the CHARGE (CHildhood Autism Risks from Genetics and Environment) study. J Autism Dev Disord 2013; 43: 25–33.
Zerbo O, Qian Y, Yoshida C, Grether JK, Van de Water J, Croen LA . Maternal infection during pregnancy and autism spectrum disorders. J Autism Dev Disord 2013; 45: 4015–4025.
Bauman MD, Iosif AM, Smith SE, Bregere C, Amaral DG, Patterson PH . Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol Psychiatry 2014; 75: 332–341.
Patterson PH . Maternal infection and autism. Brain Behav Immun 2012; 26: 393.
Smith SE, Li J, Garbett K, Mirnics K, Patterson PH . Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci 2007; 27: 10695–10702.
Gabay C, Kushner I . Acute-phase proteins and other systemic responses to inflammation. N Engl J Med 1999; 340: 448–454.
Goines PE, Croen LA, Braunschweig D, Yoshida CK, Grether J, Hansen R et al. Increased mid-gestational IFN-gamma, IL-4, and IL-5 in women giving birth to a child with autism: a case-control study. Mol Autism 2011; 2: 13.
Hage FG, Szalai AJ . C-reactive protein gene polymorphisms, C-reactive protein blood levels, and cardiovascular disease risk. J Am Coll Cardiol 2007; 50: 1115–1122.
Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 2011; 123: 731–738.
Okada Y, Takahashi A, Ohmiya H, Kumasaka N, Kamatani Y, Hosono N et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum Mol Genet 2011; 20: 1224–1231.
Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am J Hum Genet 2008; 82: 1185–1192.
Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF et al. Genetic loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA 2009; 302: 37–48.
Wensley F, Gao P, Burgess S, Kaptoge S, Di Angelantonio E, Shah T et al. Association between C reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. BMJ 2011; 342: d548.
Ernst GD, de Jonge LL, Hofman A, Lindemans J, Russcher H, Steegers EA et al. C-reactive protein levels in early pregnancy, fetal growth patterns, and the risk for neonatal complications: the Generation R Study. Am J Obstet Gynecol 2011; 205: 132.e1–132.e12.
Grgic G, Skokic F, Bogdanovic G . C-reactive protein as a biochemical marker of idiopathic preterm delivery. Med Arh 2010; 64: 132–134.
Hwang HS, Kwon JY, Kim MA, Park YW, Kim YH . Maternal serum highly sensitive C-reactive protein in normal pregnancy and pre-eclampsia. Int J Gynaecol Obstet 2007; 98: 105–109.
Lohsoonthorn V, Qiu C, Williams MA . Maternal serum C-reactive protein concentrations in early pregnancy and subsequent risk of preterm delivery. Clin Biochem 2007; 40: 330–335.
Tjoa ML, van Vugt JM, Go AT, Blankenstein MA, Oudejans CB, van Wijk IJ . Elevated C-reactive protein levels during first trimester of pregnancy are indicative of preeclampsia and intrauterine growth restriction. J Reprod Immunol 2003; 59: 29–37.
Brown AS, Sourander A, Hinkka-Yli-Salomaki S, McKeague IW, Sundvall J, Surcel HM . Elevated maternal C-reactive protein and autism in a national birth cohort. Mol Psychiatry 2014; 19: 259–264.
Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C . Prevalence of autism in a US metropolitan area. JAMA 2003; 289: 49–55.
Dickerson F, Stallings C, Origoni A, Vaughan C, Khushalani S, Yolken R . Elevated C-reactive protein and cognitive deficits in individuals with bipolar disorder. J Affect Disord 2013; 150: 456–459.
Grether JK, Croen LA, Anderson MC, Nelson KB, Yolken RH . Neonatally measured immunoglobulins and risk of autism. Autism Res 2010; 3: 323–332.
Abdallah MW, Larsen N, Grove J, Bonefeld-Jorgensen EC, Norgaard-Pedersen B, Hougaard DM et al. Neonatal chemokine levels and risk of autism spectrum disorders: findings from a Danish historic birth cohort follow-up study. Cytokine 2013; 61: 370–376.
Tsang KM, Croen LA, Torres AR, Kharrazi M, Delorenze GN, Windham GC et al. A genome-wide survey of transgenerational genetic effects in autism. PLoS One 2013; 8: e76978.
Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics 2011; 98: 79–89.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Yang J, Lee SH, Goddard ME, Visscher PM . GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82.
Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA et al. A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26: 2336–2337.
Belo L, Santos-Silva A, Rocha S, Caslake M, Cooney J, Pereira-Leite L et al. Fluctuations in C-reactive protein concentration and neutrophil activation during normal human pregnancy. Eur J Obstet Gynecol Reprod Biol 2005; 123: 46–51.
Sacks GP, Seyani L, Lavery S, Trew G . Maternal C-reactive protein levels are raised at 4 weeks gestation. Hum Reprod 2004; 19: 1025–1030.
Watts DH, Krohn MA, Wener MH, Eschenbach DA . C-reactive protein in normal pregnancy. Obstet Gynecol 1991; 77: 176–180.
Acknowledgements
The work was funded by Grant 3R01ES016669 from National Institute of Environmental Health Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zerbo, O., Traglia, M., Yoshida, C. et al. Maternal mid-pregnancy C-reactive protein and risk of autism spectrum disorders: the early markers for autism study. Transl Psychiatry 6, e783 (2016). https://doi.org/10.1038/tp.2016.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2016.46
This article is cited by
-
Sex-specific and sex-independent steroid-related biomarkers in early second trimester maternal serum associated with autism
Molecular Autism (2023)
-
A human leukocyte antigen imputation study uncovers possible genetic interplay between gut inflammatory processes and autism spectrum disorders
Translational Psychiatry (2023)
-
Maternal levels of acute phase proteins in early pregnancy and risk of autism spectrum disorders in offspring
Translational Psychiatry (2022)
-
Assessment of a combination of plasma anti-histone autoantibodies and PLA2/PE ratio as potential biomarkers to clinically predict autism spectrum disorders
Scientific Reports (2022)
-
A profile and review of findings from the Early Markers for Autism study: unique contributions from a population-based case–control study in California
Molecular Autism (2021)