Abstract
Integration of complex data and data management represent major challenges in large-scale biobank-based post-genome era research projects like GenomEUtwin (an international collaboration between eight Twin Registries) with extensive amounts of genotype and phenotype data combined from different data sources located in different countries. The challenge lies not only in data harmonization and constant update of clinical details in various locations, but also in the heterogeneity of data storage and confidentiality of sensitive health-related and genetic data. Solid infrastructure must be built to provide secure, but easily accessible and standardized, data exchange also facilitating statistical analyses of the stored data. Data collection sites desire to have full control of the accumulation of data, and at the same time the integration should facilitate effortless slicing and dicing of the data for different types of data pooling and study designs. Here we describe how we constructed a federated database infrastructure for genotype and phenotype information collected in seven European countries and Australia and connected this database setting via a network called TwinNET to guarantee effortless data exchange and pooled analyses. This federated database system offers a powerful facility for combining different types of information from multiple data sources. The system is transparent to end users and application developers, since it makes the set of federated data sources look like a single system. The user need not be aware of the format or site where the data are stored, the language or programming interface of the data source, how the data are physically stored, whether they are partitioned and/or replicated or what networking protocols are used. The user sees a single standardized interface with the desired data elements for pooled analyses.
Similar content being viewed by others
Introduction
The post-genome era provides us with technologies for collecting vast amounts of molecular information from biological samples, clinical phenotype and collected lifestyle data of individuals. The goal of many biobank-related research efforts is to link these data to data from epidemiological registers and health-care databases. A proof of principle is provided by the international GenomEUtwin project,1 (http://www.genomeutwin.org), a European Commission-funded collaboration between Twin Registries in the Netherlands, Denmark, Norway, Sweden, Finland, Italy, UK and Australia. By pooling epidemiological and phenotype information from over 600 000 twin pairs, and genotype data from an ascertained fraction of those, the collaboration aims to identify genetic variants associated with common diseases. Many of these twin cohorts include phenotype data for clinical entities, complex longitudinal, life-long data of lifestyle and environment. Furthermore, most participating twin cohorts have obtained a permit to link the study samples to national health-care registries such as the Inpatient or Hospital Discharge Registry, the Cancer Registry and the Cause of Death Registry, which makes GenomEUtwin an epidemiologic goldmine. These features make this study sample a unique resource, not only for gene identification but also as a most efficient vehicle for identification of the genetic and lifestyle/environmental risk factors causing common diseases. However, a solid database infrastructure is required for effortless data handling and integrated data analyses.
Incorporating genome-wide information into this effort requires integration of genotype and phenotype data collected over several decades in different countries. Massive data sets constructed with the information collected in different formats create massive technical challenges. Moving towards a global information infrastructure is directly connected to the issues of semantic interoperability through standardized formats and consensus terminologies.2 In spite of several large-scale projects and global achievements in standardization, the data handling issues remain an isolated and unexplored area of informatics severely hampering scientific achievements.
Traditionally, data that have been collected in studies such as GenomEUtwin are combined into one centralized repository, a data warehouse, using strict data submission protocols. This creates a large amount of rigidity in the data collection phase and also complicates the necessary constant update of the warehouse information. In the GenomEUtwin, a complementary approach was chosen where data are accessed on demand from participating centers, using direct database connections. This strategy offers flexible infrastructure for data sharing and collaboration between centers, providing the possibility to adapt the informatics infrastructure easily to different research needs.
The information system of GenomEUtwin is based on the following requirements:
-
All the locally collected phenotype and clinical data remain under the control of the national centers and unauthorized access and usage is prohibited. Security and access controls are based on policy rules approved by local authorities and all partners of this collaboration.
-
Genotype and phenotype data are stored and maintained in separate operational databases. Data can be combined and stored for pooled data analysis abiding by rules monitored by the ethical core of GenomEUtwin and approved by the steering group.
-
Developed common standards are used for all stored phenotype and genotype data.3
-
A unique randomized identifier, called GenomEUtwin identifier (EUid), is created for each subject. The EUid number consists of four parts: country, randomized number, twin identification number and a check sum.4 Each center is responsible for creating and maintaining the EUid numbers for their individuals.
Interoperability and data management
There are three steps in data integration: data are first extracted and harmonized into a common format at data provider site. In the second step, the harmonized data are transferred to a data-collecting center where it is checked and loaded into a common database (third step).
The first step, data extraction and harmonization, is often extremely time-consuming because, owing to differences in the underlying study designs and annotations, data cannot be directly mapped into a common consensus format. The second step, the data transfer, is equally critical and it defines the flexibility of the data integration system. Traditionally, data have been sent to a data-collecting center, where they are decrypted, checked and loaded into a central database. The process is often slow owing to long communication delays. This approach has been used, for example, in UK Biobank5 and MONICA.6
For the TwinNET, an alternative approach was chosen where data are loaded directly at the data provider site and made available using database federation.7, 8 Benefit of the approach is that data can be made available faster since data management work is distributed and done by most experienced personnel. There is also the possibility to quickly explore new unharmonized data sets, which can be copied from productions systems using SQL statements. This is important in study planning and ad hoc data analysis. Another important benefit of database federation is that the data provider can retain control over the data and make it available as needed.
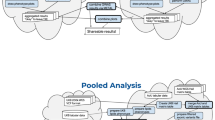
The concept of a database federation is not new. It has been available on relational database management systems for over two decades.9 In a federated system, remote data tables or data objects in general are made available through an integrating database using special database views (Figure 1), which are like local view tables that can be joined in SQL queries with other tables and views.
A federated database system is a type of database management system that transparently integrates multiple autonomous database systems into a single federated database. The constituent databases are interconnected via a computer network, and may be geographically decentralized. A federated database (or virtual database) is the fully integrated, logical composite of all constituent databases in a federated database system. Data sources could be both structured (relational databases, XML documents, etc) and/or unstructured (Excel spread sheets, medical records, etc). Because various database management systems employ different query languages, federated database systems can apply ‘wrappers’ to the subqueries to translate them into the appropriate query language.
Implementation
Connecting the data collection sites
The network architecture of TwinNET is Hub-and-Spoke, where the Hub is the integration node and Spokes are data-providing centers, for example, the twin registries (Figure 2). To maximize security, all unneeded connections and network protocols are disabled and centers can only connect to the Hub. Connections are made secure using virtual private network (VPN) tunnels,10, 11 which are initiated from the data-providing centers.
General topology of TwinNET. Twin registries are the data providers, which are connected to a Hub using direct database connections. The Hub provides a single database access point for the data using the DB2 and Discovery Link (WebSphere Federation Server). Federated data forms remote databases that can be shared through the DB2 database management system as any other data stored locally in a relational database.
Database servers at the data-providing centers are maintained according to agreed security policies.12 The server is located in the TwinNET demilitarized zone (DMZ) (Figures 3 and 4) and it can be disconnected from the local area network to simplify security management. The local database, the TwinMART, is updated by copying data from production databases using transient connections according to local security rules. Users can access data from the Hub using a Web interface and terminal services provided by the Genome Informatics Unit,13 which also hosts the computing services. It is also possible to host TwinMART servers themselves. This kind of hosting service should be easy to implement using preconfigured virtual machines,14 which can be copied for new partners.
The minimum requirement for centers to join the TwinNET network is to have VPN gateway which meets agreed security policy requirements. The database server, which is in demilitarized zone (DMZ), is a running relational database management system supported by the center. The server is connected to the Hub over Internet using appropriate database protocol that is tunneled through established VPN connections. Implementation is independent of tunneling technique. In current implementation we have used Cisco compatible IPSec protocol7 and SSL VPN protocol as implemented in the OpenVPN software.8
Remote databases are linked to the DB2 relational database management system instance running on the Hub. Remote tables are mounted into the DB2 database using Discovery Link7 extensions that provide the so-called wrappers for mapping tables and data types from different vendors.
The DB2 and Discovery Link bundle, together called the WebSphere Federation Server, was chosen because it provides an extensive number of wrappers for different relational and also non-relational data sources commonly used in life science research. These remote objects can be transparently cached and queried using dynamically optimized SQL.7 The WebSphere Federation Server also provides configurable mappings between other schema objects such as functions and user accounts that simplify management of data sources. The WebSphere Federation Server runs on different operating systems and integrates with open source development work through free products like Java/JDBC15 and Eclipse.16
Connecting the genome and phenome data from multiple sites
Genotype data, already generated in earlier studies or constantly produced by genotyping centers without appropriate database backing, are maintained and collected by the centralized genotype data collection site (in this particular case of GenomEUtwin at the Finnish Genome Center at the University of Helsinki). Data management is handled inside a local area network where it is checked and harmonized. Approved data are then replicated into a server located in the TwinNET zone, from where they can be accessed by the Hub. Other genotyping centers can join the study provided they present satisfactory database and quality-control services. In the setting of GenomEUtwin, the second genotyping center joined via the TwinNET is the Department of Molecular Medicine at the University of Uppsala, Sweden.
Phenotype data are provided directly by the twin centers. The data are first harmonized at the center and loaded into a local database server located on the TwinNET zone. The phenotype data are more diverse than genotype data, owing to varying ambitions throughout the past 40 years of data collection in some twin cohorts.17 Predefined database schemas are implemented for major modern relational database management systems to simplify the data-loading process. Schemas are designed to be as simple as possible, securing the minimum amount of information needed for specific studies. Besides simplicity, another goal is to define most data constraints at the database level. This ensures the detection of possible errors already at the loading time and enables correction by personnel most experienced with the data. All the database activities are supervised and coordinated by the database core to ensure that work is not repeated unnecessarily and predefined policies are followed carefully.12
Standardization of the data
Common standards are a key to data integration. A unique, randomized identifier is established for each subject. The genotype database complies with the conceptual data model made for Polymorphism Markup Language.18 This specification facilitates data integration with other studies and databases. Phenotype data are standardized and harmonized within each project or special study. Within the GenomEUtwin, the data specifications have been created for weight, height and BMI, for migraine with both questionnaire data and details of clinical phenotype, and for serum lipid values, insulin and glucose content and other measures of metabolic traits and for cardiovascular disease studies. The databases currently contain integrated information for the initial test traits, weight and height (and BMI) from more than 250 000 individuals, for migraine questionnaire and details of clinical phenotype from 8000 individuals, for serum lipid values and insulin and glucose content and other measures of metabolic traits for over 20 000 individuals. Data harmonization for numerous parameters of cardiovascular traits is in process.
Similarly, the genotyping sites have produced rigorous QC systems and all the genotypes and alleles are harmonized across two genotyping sites (Helsinki and Uppsala), and the quality controls are rigorously monitored before the data are loaded in the genome database. The accessible genotypes in the database are more than 20 million at present and the number is growing at an accelerating speed.
Data security
Data must be stored so that unauthorized access is prohibited. All databases and data sets maintained under the TwinNET are anonymous – they contain no identifiers that can be used to identify individuals in the studies. The only allowed identifiers that can be associated with subject or samples are randomized GenomEUtwin identifiers. Further, genome and phenome data are stored in separate operational databases. The data can only be combined and stored in one place for analysis purposes, under the TwinNET policy rules.12
Discussion
In the current era of numerous biobank-based research efforts, data collection, storage and integration present massive challenges. Genetic profiles for thousands or tens of thousands of individuals are combined with the detailed clinical and epidemiological data sets, often longitudinal, which are continuously accumulating and updated with the information from multiple registries. The GenomEUtwin project is an example of a large international research program that aims to harmonize and integrate data from already existing and newly collected studies across Europe and Australia. Our federated database was designed as a tool to facilitate searching, updating and managing collected information obtained from various and diverse data sources. A federated database system facilitates, except data storage and update, pooled analyses across individual studies and data collection centers. This system, the TwinNET, provides a transparent virtual view of data stored in various scientific computer systems and databases. The concept is drastically different from more conventional solutions where all data are periodically extracted to a large central data warehouse. Advantages of this solution include the ability to utilize computational resources provided by the individual centers, and the possibility to use modern dynamic query optimization techniques for improved scalability and performance in communication and access. To our knowledge, this is the first effort to integrate massive longitudinal data sets into one database with harmonized content and easy access for all involved investigators. Within this setting, the twin investigators communicate effortlessly, and also the valuable twin cohort data, collected and updated over the past 40 years are stored in a systematic, harmonized way making the effort less dependent on individual data collectors. This significantly increases the accumulated value of the data and improves accessibility of this valuable dataset to the international scientific community.
Any computer-dependent system has a tendency to become increasingly complex and outdated, considering the continuous development of software and hardware. As the amount of software increases, so does the number of possible sources of error. We have made an effort to build the TwinNET as simple as possible using standard technologies. The implementation is not tied to any specific platform from the end users' and application developers' points of view and there is no need to implement and manage separate data service layers. All data, whether from remote or local sources, can be accessed immediately using standard query language and database protocols.
One major weakness of the prospective cohort approach is the enormous amount of time and money that must be invested before information can be retrieved. Both genome and phenome information evolve continuously and the number of individual samples with massive amount of data is rapidly increasing in databases. Consequently, new information sources (possibly other European cohorts) will need to be added continuously to the TwinNET environment. The more information is made available, the more important it is to provide a scalable infrastructure to grow with the increasing data volume and complexity. The federated approach described here aims to achieve such scalability, making it flexible and increasing its life length. With the TwinNET project, we have demonstrated that seven European countries and Australia can share complex phenotype/genotype data. Some of the twin cohorts started collecting their data in the late 1950s while others have just started to collect data. Nevertheless, using the federated approach, we have ‘connected’ the existing information of 600 000 twins in Europe. This project has the potential to revolutionize both epidemiologic and clinical research and will pave the way for incorporating other large population cohorts and biobanks.
References
Peltonen L : GenomEUtwin: a strategy to identify genetic influences on health and disease. Twin Res 2003; 6: 354–360.
Berners-Lee T, Hendler J, Lassila O : The Semantic Web. Sci Am 2001; 284: 34–43.
Björklund A, Litton J-E : Data Format and Variable Standard for GenomEUtwin's Phenotype Database Prototype, Version: 4.0 2005.
Litton J-E, Muilu J, Björklund A, Leinonen A, Pedersen NL : Data modeling and communication in GenomEUtwin. Twin Res 2003; 6: 383–390.
Ollier W, Sprosen T, Peakman T : UK Biobank: from concept to reality. Pharmacogenomics 2005; 639–646.
Tunstall-Pedoe H, Kuulasmaa K, Amouyel P, Arveiler D, Rajakangas AM, Pajak A : Myocardial infarction and coronary deaths in the World Health Organization MONICA Project. Registration procedures, event rates, and case-fatality rates in 38 populations from 21 countries in four continents. Circulation 1984; 90: 583–612.
Haas LM, Lin ET, Roth MA : Data integration through database federation. IBM Syst J 2002; 41: 580–596.
Bourbonnais S, Gogate VM, Haas LM et al: Towards an information infrastructure for the grid. IBM Syst J 2004; 43: 665–688.
Gomer T, Thompson GR, Chung CW, Barkmeyer E, Carter F, Templeton M : Heterogeneous distributed database systems for production use. ACM Comput Surv 1990; 22: 237–266.
Herscovitz E : Secure virtual private networks: the future of data communications. Int J Network Mgmt 1999; 9: 213–220.
Johansen JR, Litton JE : Security Policies for TwinNET 2005, Version 1.4 http://www.genomeutwin.org/Member/mmain.htm.
Barham P, Dragovic B, Fraser K et al: Xen and the art of virtualization. Proceedings of the Nineteenth ACM Symposium on Operating Systems Principles, pp 2003; 164–177.
Lichtenstein P, De faire U, Floderus B, Svartengren M, Svedberg P, Pedersen NL : The Swedish Twin Registry: a unique resource for clinical epidemiological and genetic studies. J Intern Med 2002; 252: 184–205.
Acknowledgements
This project was supported by the European Commission under the program ‘Quality of Life and Management of the Living Resources’ of 5th Framework Program (no. QLG2-CT-2002-01254). We would like to thank the twins in all the countries for their participation. The following Universities/Hospitals are included in the TwinNET: The Netherlands Twin Registry, Department of Biological Psychology, Vrije University, Amsterdam; Department of Molecular Medicine, National Public Health Institute, Helsinki, Finland; Finnish Twin Cohort Study, University of Helsinki, Finland; Finnish Genome Center, University of Helsinki, Finland; Department of Medical Sciences, Uppsala University, Sweden; Swedish Twin Registry, Karolinska Institutet, Stockholm, Sweden; Danish Twin Registry, University of Southern Denmark, Odense, Denmark; Italian Twin Registry, Instituto Superiore di Sanità, Rome, Italy; Norwegian Twin Registry, The Norwegian Institute of Public Health, Oslo, Norway; Department of Epidemiology and Public Health, University of Belfast, UK; Leiden University Medical Centre, Leiden, the Netherlands; Twin Research and Genetic Epidemiology Unit, St Thomas' Hospital, London, UK; Australian Twin Registry, Queensland Institute of Medical Research, Brisbane, Australia. Furthermore, this work could not have been carried out without the members in the GenomEUtwin Database Core and GenomEUtwin Security Group: Ann Björklund, Timo Miettinen, Anne Leinonen, Emad Qweitin, Kari Kuulasmaa, Markus Perola, Leena Peltonen, Eugenio Carrani, HJ van der Wijk, Harry Beeby, Roberto Aielli, Rodolfo Cotichini, Nieuwboer RT, Gonneke Willemsen, Patrik Magnusson, Juri Ahokas, Fons Ullings, Tatjana Dukic, Juha Saharinen, Jaason Haapakoski, Gunnar Petersson, Jennifer Harris, Jenny Carlsson, Johan Söderberg, Jon Johansen, Ingunn Brandt, Kauko Heikkilä , Lars Hvidberg, Mats Jonsson, Matti Siivola, Maurice Michiels, Axel Skytthe, Zygimantas Cepaitis, Pontus Lindqvist and George Ölund.
We would also like to acknowledge Jean-Christophe Mestres, Edith Lefrançois and Jean-Luc Collet from the IBM Research Center at LaGaude, France.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Muilu, J., Peltonen, L. & Litton, JE. The federated database – a basis for biobank-based post-genome studies, integrating phenome and genome data from 600 000 twin pairs in Europe. Eur J Hum Genet 15, 718–723 (2007). https://doi.org/10.1038/sj.ejhg.5201850
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201850
Keywords
This article is cited by
-
The governance structure for data access in the DIRECT consortium: an innovative medicines initiative (IMI) project
Life Sciences, Society and Policy (2018)
-
Data harmonization and federated analysis of population-based studies: the BioSHaRE project
Emerging Themes in Epidemiology (2013)
-
Linkage of Data from Diverse Data Sources (LDS): A Data Combination Model Provides Clinical Data of Corresponding Specimens in Biobanking Information System
Journal of Medical Systems (2013)
-
The International Collaboration for Autism Registry Epidemiology (iCARE): Multinational Registry-Based Investigations of Autism Risk Factors and Trends
Journal of Autism and Developmental Disorders (2013)