Abstract
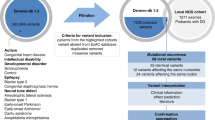
We combined de novo mutation (DNM) data from 10,927 individuals with developmental delay and autism to identify 253 candidate neurodevelopmental disease genes with an excess of missense and/or likely gene-disruptive (LGD) mutations. Of these genes, 124 reach exome-wide significance (P < 5 × 10−7) for DNM. Intersecting these results with copy number variation (CNV) morbidity data shows an enrichment for genomic disorder regions (30/253, likelihood ratio (LR) +1.85, P = 0.0017). We identify genes with an excess of missense DNMs overlapping deletion syndromes (for example, KIF1A and the 2q37 deletion) as well as duplication syndromes, such as recurrent MAPK3 missense mutations within the chromosome 16p11.2 duplication, recurrent CHD4 missense DNMs in the 12p13 duplication region, and recurrent WDFY4 missense DNMs in the 10q11.23 duplication region. Network analyses of genes showing an excess of DNMs highlights functional networks, including cell-specific enrichments in the D1+ and D2+ spiny neurons of the striatum.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All variant data in this study are available to download from denovo-db v.1.5 (http://denovo-db.gs.washington.edu/). Human MTG single-nucleus RNA-seq data and clusters can be downloaded from the Allen Institute for Brain Science website at http://celltypes.brain-map.org/download.
References
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005).
Tuzun, E. et al. Fine-scale structural variation of the human genome. Nat. Genet. 37, 727–732 (2005).
de Vries, B. B. et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 77, 606–616 (2005).
Bailey, J. A., Yavor, A. M., Massa, H. F., Trask, B. J. & Eichler, E. E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017 (2001).
de Vries, B. B. et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J. Med. Genet. 38, 145–150 (2001).
Firth, H. V. & Wright, C. F. The Deciphering Developmental Disorders (DDD) study. Dev. Med. Child Neurol. 53, 702–703 (2011).
O’Roak, B. J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015).
Stessman, H. A. et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515–526 (2017).
O’Roak, B. J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Samocha, K. E. et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944–950 (2014).
Turner, T. N. et al. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 24, 5995–6002 (2015).
Geisheker, M. R. et al. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 20, 1043–1051 (2017).
Lelieveld, S. H. et al. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 101, 478–484 (2017).
Cooper, G. M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Kaminsky, E. B. et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 13, 777–784 (2011).
Coe, B. P. et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46, 1063–1071 (2014).
Sanders, S. J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
Turner, T. N. et al. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 45, D804–D811 (2017).
Matson, J. L. & Shoemaker, M. Intellectual disability and its relationship to autism spectrum disorders. Res. Dev. Disabil. 30, 1107–1114 (2009).
American Psychiatric Association Diagnostic and statistical manual of mental disorders, 5th edition: (DSM−5) (APA Publishing, Arlington, 2013).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Halvardson, J. et al. Mutations in HECW2 are associated with intellectual disability and epilepsy. J. Med. Genet. 53, 697–704 (2016).
Hashimoto, R. et al. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 61, 199–206 (2016).
Krumm, N. et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015).
Lee, H., Lin, M. C., Kornblum, H. I., Papazian, D. M. & Nelson, S. F. Exome sequencing identifies de novo gain of function missense mutation in KCND2 in identical twins with autism and seizures that slows potassium channel inactivation. Hum. Mol. Genet. 23, 3481–3489 (2014).
Lelieveld, S. H. et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 19, 1194–1196 (2016).
Michaelson, J. J. et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012).
Moreno-Ramos, O. A., Olivares, A. M., Haider, N. B., de Autismo, L. C. & Lattig, M. C. Whole-exome sequencing in a South American cohort links ALDH1A3, FOXN1 and retinoic acid regulation pathways to autism spectrum disorders. PLoS ONE. 10, e0135927 (2015).
Rauch, A. et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682 (2012).
RK, C. Y. et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611 (2017).
Tavassoli, T. et al. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med. Genet. 15, 35 (2014).
Turner, T. N. et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 98, 58–74 (2016).
Yuen, R. K. et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 1, 160271–1602710 (2016).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Wang, T. et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 7, 13316 (2016).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Le Meur, N. et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J. Med. Genet. 47, 22–29 (2010).
Hormozdiari, F., Penn, O., Borenstein, E. & Eichler, E. E. The discovery of integrated gene networks for autism and related disorders. Genome Res. 25, 142–154 (2015).
Endele, S. et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 42, 1021–1026 (2010).
Ching, M. S. et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 937–947 (2010).
Stephenson, J. R. et al. A novel human CAMK2A mutation disrupts dendritic morphology and synaptic transmission, and causes ASD-related behaviors. J. Neurosci. 37, 2216–2233 (2017).
Dougherty, J. D., Schmidt, E. F., Nakajima, M. & Heintz, N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 38, 4218–4230 (2010).
Xu, X., Wells, A. B., O’Brien, D. R., Nehorai, A. & Dougherty, J. D. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 34, 1420–1431 (2014).
Deshpande, A. & Weiss, L. A. Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Dev. Neurobiol. 78, 519–530 (2018).
Koolen, D. A. et al. The Koolen-de Vries syndrome: a phenotypic comparison of patients with a 17q21.31 microdeletion versus a KANSL1 sequence variant. Eur. J. Hum. Genet. 24, 652–659 (2016).
Phelan, K. & Rogers, R. C. Phelan-McDermid Syndrome. in GeneReviews(R) (eds. Adam, M. P. et al.) (Seattle (WA), 1993).
Bi, W. et al. Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum. Genet. 115, 515–524 (2004).
Han, J. Y. et al. Identification of a novel de novo nonsense mutation of the NSD1 gene in monozygotic twins discordant for Sotos syndrome. Clin. Chim. Acta 470, 31–35 (2017).
Izumi, K. et al. Interstitial microdeletion of 4p16.3: contribution of WHSC1 haploinsufficiency to the pathogenesis of developmental delay in Wolf-Hirschhorn syndrome. Am. J. Med. Genet. A 152A, 1028–1032 (2010).
Shimbo, H. et al. Haploinsufficiency of BCL11A associated with cerebellar abnormalities in 2p15p16.1 deletion syndrome. Mol. Genet. Genomic Med. 5, 429–437 (2017).
Kleefstra, T. et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J. Med. Genet. 46, 598–606 (2009).
Fergelot, P. et al. Phenotype and genotype in 52 patients with Rubinstein-Taybi syndrome caused by EP300 mutations. Am. J. Med. Genet. A. 170, 3069–3082 (2016).
Kumar, R. A. et al. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 17, 628–638 (2008).
Labonne, J. D. et al. A microdeletion encompassing PHF21A in an individual with global developmental delay and craniofacial anomalies. Am. J. Med. Genet. A 167A, 3011–3018 (2015).
McCool, C., Spinks-Franklin, A., Noroski, L. M. & Potocki, L. Potocki-Shaffer syndrome in a child without intellectual disability-The role of PHF21A in cognitive function. Am. J. Med. Genet. A 173, 716–720 (2017).
Leroy, C. et al. The 2q37-deletion syndrome: an update of the clinical spectrum including overweight, brachydactyly and behavioural features in 14 new patients. Eur. J. Hum. Genet. 21, 602–612 (2013).
Klebe, S. et al. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur. J. Hum. Genet. 20, 645–649 (2012).
Halder, D. et al. Suppression of Sin3A activity promotes differentiation of pluripotent cells into functional neurons. Sci. Rep. 7, 44818 (2017).
Witteveen, J. S. et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat. Genet. 48, 877–887 (2016).
Amir, R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999).
Jansen, S. et al. De novo truncating mutations in the last and penultimate exons of PPM1D cause an intellectual disability syndrome. Am. J. Hum. Genet. 100, 650–658 (2017).
DeMari, J. et al. CLTC as a clinically novel gene associated with multiple malformations and developmental delay. Am. J. Med. Genet. A 170A, 958–966 (2016).
Fusco, C. et al. Smaller and larger deletions of the Williams Beuren syndrome region implicate genes involved in mild facial phenotype, epilepsy and autistic traits. Eur. J. Hum. Genet. 22, 64–70 (2014).
Buxbaum, J. D. et al. Association between a GABRB3 polymorphism and autism. Mol. Psychiatry 7, 311–316 (2002).
Guella, I. et al. De novo mutations in YWHAG cause early-onset epilepsy. Am. J. Hum. Genet. 101, 300–310 (2017).
Asadollahi, R. et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J. Med. Genet. 51, 677–688 (2014).
Harrington, A. J. et al. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. eLife 5, e20059 (2016).
Paciorkowski, A. R. et al. MEF2C haploinsufficiency features consistent hyperkinesis, variable epilepsy, and has a role in dorsal and ventral neuronal developmental pathways. Neurogenetics 14, 99–111 (2013).
Kohannim, O. et al. Discovery and replication of gene influences on brain structure using LASSO regression. Front. Neurosci. 6, 115 (2012).
Weiss, K. et al. De novo mutations in CHD4, an ATP-dependent chromatin remodeler gene, cause an intellectual disability syndrome with distinctive dysmorphisms. Am. J. Hum. Genet. 99, 934–941 (2016).
Berko, E. R. et al. De novo missense variants in HECW2 are associated with neurodevelopmental delay and hypotonia. J. Med. Genet. 54, 84–86 (2017).
Harripaul, R. et al. Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry 23, 973−984 (2018).
Wang, Q., Moore, M. J., Adelmant, G., Marto, J. A. & Silver, P. A. PQBP1, a factor linked to intellectual disability, affects alternative splicing associated with neurite outgrowth. Genes Dev. 27, 615–626 (2013).
Levy, J. et al. Molecular and clinical delineation of 2p15p16.1 microdeletion syndrome. Am. J. Med. Genet. A 173, 2081–2087 (2017).
Dheedene, A., Maes, M., Vergult, S. & Menten, B. A de novo POU3F3 deletion in a boy with intellectual disability and dysmorphic features. Mol. Syndromol. 5, 32–35 (2014).
Carlston, C. M. et al. Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring-Opitz Syndrome. Hum. Mutat. 38, 517–523 (2017).
He, X. et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 9, e1003671 (2013).
Werling D. M. et al. Limited contribution of rare, noncoding variation to autism spectrum disorder from sequencing of 2,076 genomes in quartet families. BioRxiv https://dx.doi.org/10.1101/127043 (2017).
Turner T. N. et al. Genomic patterns of de novo mutation in simplex autism. Cell 171, 710–722.e12 (2017).
Park, S. M., Park, H. R. & Lee, J. H. MAPK3 at the autism-linked human 16p11.2 locus influences precise synaptic target selection at drosophila larval neuromuscular junctions. Mol. Cells 40, 151–161 (2017).
Pucilowska, J. et al. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J. Neurosci. 35, 3190–3200 (2015).
Blizinsky, K. D. et al. Reversal of dendritic phenotypes in 16p11.2 microduplication mouse model neurons by pharmacological targeting of a network hub. Proc. Natl Acad. Sci. USA 113, 8520–8525 (2016).
Langen, M. et al. Changes in the development of striatum are involved in repetitive behavior in autism. Biol. Psychiatry 76, 405–411 (2014).
Platt, R. J. et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 19, 335–350 (2017).
Reim, D. et al. Proteomic analysis of post-synaptic density fractions from Shank3 Mutant mice reveals brain region specific changes relevant to autism spectrum disorder. Front. Mol. Neurosci. https://doi.org/10.3389/fnmol.2017.00026 (2017).
Balsters, J. H., Mantini, D. & Wenderoth, N. Connectivity-based parcellation reveals distinct cortico-striatal connectivity fingerprints in autism spectrum disorder. Neuroimage 170, 412–423 (2018).
Shohat, S., Ben-David, E. & Shifman, S. Varying intolerance of gene pathways to mutational classes explain genetic convergence across neuropsychiatric disorders. Cell Rep. 18, 2217–2227 (2017).
Kaya, N. et al. KCNA4 deficiency leads to a syndrome of abnormal striatum, congenital cataract and intellectual disability. J. Med. Genet. 53, 786–792 (2016).
Flanigan, M. & LeClair, K. Shared motivational functions of ventral striatum D1 and D2 medium spiny neurons. J. Neurosci. 37, 6177–6179 (2017).
Sanders, S. J. First glimpses of the neurobiology of autism spectrum disorder. Curr. Opin. Genet. Dev. 33, 80–92 (2015).
Schreiweis, C. et al. Humanized Foxp2 accelerates learning by enhancing transitions from declarative to procedural performance. Proc. Natl Acad. Sci. USA 111, 14253–14258 (2014).
Chen, Y. C. et al. Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat. Neurosci. 19, 1513–1522 (2016).
Stessman, H. A., Bernier, R. & Eichler, E. E. A genotype-first approach to defining the subtypes of a complex disease. Cell 156, 872–877 (2014).
Bernier, R. et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014).
Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220 (2010).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Acknowledgements
We wish to thank T. Turner and J. Shendure for helpful discussion and T. Brown for edits. This research was supported, in part, by the following: the Simons Foundation Autism Research Initiative (SFARI 303241) and US National Institutes of Health (NIH R01MH101221) to E.E.E. The J.D.D. laboratory is supported by a NARSAD Independent Investigator Award from the Brain and Behavior Research Foundation and NIH grant 5R01MH107515-03. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). E.E.E. is supported by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
B.P.C. and E.E.E. designed the study. B.P.C. performed the primary statistical data analysis. B.P.C., H.A.F.S., and M.R.G. curated DNMs and performed enrichment analyses. A.S. assisted with statistical analyses and interpretation. R.A.B. performed phenotype analysis. T.E.B. and E.S.L. performed the human expression analysis. A.M.L. and J.D.D. performed CSEA on cortex and assisted with additional CSEA and TSEA. F.H. performed the gene network analysis. B.P.C. and E.E.E. wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.E.E. is on the scientific advisory board of DNAnexus, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Comparison of de novo variation rates in ASD and ID/DD.
a,b, The plots compare DNM rates for genes for patients from ASD (n = 5,624 independent samples) and ID/DD (n = 5,303 independent samples) studies included in our combined analysis. More than 75% of genes show DNM in both ASD and DD patients. We identify four LGD genes (ARID1B, ANDKRD11, KMT2A, DDX3X) (a) and one missense gene (KCNQ2) (b) that are biased for an ID/DD diagnosis at a q-value threshold of 0.1 (one-tailed Fisher’s exact test). Additional candidates for phenotypic bias at nominal significance (dashed lines at P = 0.05, one-tailed Fisher’s exact test) were also identified. Larger cohorts will be needed to confirm gene biases, especially with respect to ASD.
Supplementary Figure 2 CSEA identifies bias to specific brain regions.
Cell-specific enrichment analyses (CSEA) of the union set (n = 253 independent genes) highlight a strong bias to various developing parts of the brain (color corresponds to FDR-adjusted one-tailed Fisher’s exact test P values; shaded regions closer to the center of each hexagon indicate increasing tissue specificity). a, We observe enrichment for both classes of striatal medium spiny neurons for our gene set. This tissue has been previously implicated in autism and candidate neurodevelopmental genes (J. Neurosci 34, 1420–1431, 2014), and we now observe cell-specific enrichment among genes with a significant excess of DNM. b, Application of CSEA on n = 253 independent genes to the additional cell types profiled in Zeisel et al. (Science 347, 1138–1142, 2015) identifies pyramidal neurons in layer 5 of the cortex and hippocampus. Color corresponds to FDR-adjusted one-tailed Fisher’s exact test P values; shaded regions closer to the center of each hexagon indicate increasing tissue specificity.
Supplementary Figure 3 Pan-neuronal expression patterns of candidate NDD genes.
a–c, Heatmaps demonstrating a broad pattern of inhibitory and excitatory neuronal expression (median log2 (CPM + 1)) in the NDD gene sets compared to control genes. The FWER union set shows even greater pan-neuronal-enriched expression than the larger union gene set. Rows represent individual genes and are ordered by the number of clusters with expression (median CPM > 1), and columns represent 41 inhibitory neuronal, 24 excitatory neuronal, and 6 glial clusters. d–f, Genes enriched for DNM are more broadly expressed in inhibitory (d) and excitatory (e) neurons, while genes enriched for LGD events specifically are enriched in glial expression (f). g, Comparison of control and test gene lists demonstrates similar maximum average expression (CPM) across cell types. h, Cell type specificity as measured by a beta marker score (Methods) is also similar for NDD and control genes.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Note
Supplementary Tables
Supplementary Tables 1–8
Rights and permissions
About this article
Cite this article
Coe, B.P., Stessman, H.A.F., Sulovari, A. et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet 51, 106–116 (2019). https://doi.org/10.1038/s41588-018-0288-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-018-0288-4
This article is cited by
-
Heterozygous truncating variant of TAOK1 in a boy with periventricular nodular heterotopia: a case report and literature review of TAOK1-related neurodevelopmental disorders
BMC Medical Genomics (2024)
-
An unsupervised deep learning framework for predicting human essential genes from population and functional genomic data
BMC Bioinformatics (2023)
-
Acorn: an R package for de novo variant analysis
BMC Bioinformatics (2023)
-
Astrocytic β-catenin signaling via TCF7L2 regulates synapse development and social behavior
Molecular Psychiatry (2023)
-
Methyl-CpG binding domain 2 (Mbd2) is an epigenetic regulator of autism-risk genes and cognition
Translational Psychiatry (2023)