Abstract
It has been reported that there are differences in effects on irinotecan-induced adverse reactions between UGT1A1*6 and UGT1A1*28. In order to compare those differences in the Japanese population, we examined the associations between UGT1A1 and irinotecan-induced adverse reactions using the BioBank Japan Project database. We genotyped UTG1A1*6 and UGT1A1*28 and conducted case–control analyses. A total of 651 patients (102 cases and 549 tolerant controls) were included in this study. The results showed that UGT1A1*6/*6 is a predictor of adverse drug reactions (ADRs) (p-value 0.00070, odds ratio 6.59, 95% confidence interval 2.33–18.6), whereas UGT1A1*6/*28 and UGT1A1*28/*28 were not. The subanalysis comprising only patients with UGT1A1*6/*6, UGT1A1*6/*28, and UGT1A1*28/*28 revealed a trend towards an increased risk of ADRs in patients with UGT1A1*6 (p-value 0.0092, odds ratio 4.39, 95% confidence interval 1.57–14.9). Multiple logistic regression analyses showed that use of platinum-based antineoplastic drugs and presence of UGT1A1*6/*6 were independent variables, significantly associated with ADRs. The diagnostic performance of a predictive model had a sensitivity of 49.0%, specificity of 70.1%, and a number needed to screen of 5.8. We concluded that UGT1A1 testing could be useful to predict irinotecan-induced ADRs, and that UTG1A1*6 rather than UGT1A1*28 contributed to ADR occurrence.
Similar content being viewed by others
Introduction
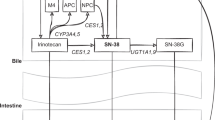
Irinotecan is widely used in Japan to treat solid tumors such as those in lung, colorectal, gastric, gynecological, and other types of cancer [1]. Adverse reactions due to irinotecan are well known, and include bone marrow suppression, diarrhea, nausea, vomiting, and others [1, 2]. Irinotecan is a prodrug, which is converted to an active metabolite, SN-38, that undergoes detoxification by uridine diphosphate glucuronosyltransferase (UGT) 1A1 to form inactive SN-38 glucuronide. Therefore, UGT1A1 alleles with decreased activity could be associated with higher plasma concentrations of SN-38 and risks of adverse drug reactions (ADRs). In Japan, pharmacogenomic testing of UGT1A1*6 and *28 has been covered by the national health insurance since 2008. Since then, it has become routine practice in Japan for patients to be tested before starting irinotecan therapy. In contrast, outside of Japan the situation is different. Our recent survey of pharmacogenomic testing of health insurance coverage revealed that UGT1A1 testing is rarely covered by the major health insurers in the US because of insufficient evidence to support its clinical utility, based on their medical policies [3].
The gap between the current situation in Japan and overseas prompted us to examine the associations of irinotecan-induced ADRs with UGT1A1*6 and *28 in the Japanese population. A number of studies examining these associations have been conducted, but the sample sizes were small in almost all the studies. In addition, systematic reviews and meta-analyses have also been performed for these associations; however, a limited number of Japanese studies were included in the studies on Asian subgroups. Consequently, the conclusions of these systematic reviews and meta-analyses remain unclear for the Japanese population. Zhang et al. successfully showed positive associations for the Japanese population; however, they only evaluated the association of neutropenia with UGT1A1*6 [4]. However, Chen et al. examined the association of neutropenia with both UGT1A1*6 and *28, showing no significant associations in Asians, which included four original Japanese studies [5]. Multiple other systematic reviews and meta-analyses, including an umbrella review, were conducted, which included studies on the Japanese population [6,7,8,9,10,11]. Some of the studies, wherein no separate analyses were performed on the Japanese population, showed positive associations in Asians; however, most studies showed no significant associations in the Japanese population [6, 7, 10, 11]. In addition, Chen et al. reported no significant associations in the Asian populations including the Japanese population for UGT1A1*28. Moreover, a study by Liu et al. showed positive associations in Asians, but all original studies in the Japanese population showed a lack of significant associations [8, 9].
The aim of this study was to evaluate an association analysis using the BioBank Japan Project database, comprising clinical and genotyping data of ~200,000 patients, for 47 different diseases, including cancers, from 12 cooperative medical institutes located across Japan [12, 13]. We performed this study to clearly assess the actual relationships between UGT1A1*6 and UGT1A1*28 with the risk of toxicities in the Japanese population using relatively large sample sizes, to separately evaluate the associations of both UGT1A1*6 or UGT1A1*28, and accumulate more evidence to show associations of ADRs due to irinotecan with UGT1A1.
Materials and methods
Subjects
All patients recruited for the BioBank Japan Project provided written informed consent, and the study protocol of this project was approved by the ethics committee at the Institute of Medical Sciences, The University of Tokyo (Tokyo, Japan), and the RIKEN Center for Integrative Medical Sciences (Yokohama, Japan). Data regarding ADRs between April 2003 and March 2018 were retrieved from the medical records of BioBank Japan-affiliated hospitals. We conducted the case–control analysis by comparing the genetic information obtained from cases that developed grade 3–5 ADRs and from those who did not develop ADRs or developed grade 1 or 2 ADRs while on irinotecan (tolerant controls). The toxicity grade was classified following the US National Cancer Institute’s Common Toxicity Criteria version 2.0 [14].
Genotyping of UGT1A1*6 and UGT1A1*28
We genotyped DNA samples of cases and tolerant controls for UGT1A1*6 and UGT1A1*28, which are commonly found and could explain the risk of ADR development due to irinotecan in the Japanese population [15,16,17,18].
For UGT1A1*6, we genotyped the subjects for rs4148323 (211G>A) using PCR, which was followed by an Invader assay. The detailed methodological protocol has been previously described by Ohnishi et al. [19]. Briefly, PCR was performed with 10 ng of genomic DNA in a total reaction volume of 20 μl using Ex Taq HS DNA polymerase (5 U/μl) (Takara Bio, Otsu, Japan). PCR was performed with an initial denaturation at 94 °C for 5 min, followed by 37 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 2 min, and a final extension at 72 °C for 5 min using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). After PCRs, the product was diluted tenfold and used as a template for the Invader assay using ABI PRISM 7900 (Applied Biosystems) according to the protocol recommended by Hologic (Marlborough, MA, USA).
For UGT1A1*28, 20 μl of the reaction mixture was used for performing PCR on each sample using the same conditions of initial denaturation and final extension as for UGT1A1*6, but the initial denaturation was followed by 40 cycles at 94 °C for 15 s, 58 °C for 45 s, and 72 °C for 1 min. After purification on MultiScreen HTS 96-well filter plates (Merck Millipore, MA, USA), the PCR products were used for sequencing using BigDye Terminator v3.1 (Applied Biosystems), 5× Big Dye Sequence Buffer (Applied Biosystems), and primers. The reaction was initiated by incubation at 96 °C for 1 min, followed by 25 cycles at 96 °C for 10 s, 55 °C for 5 s, and 60 °C for 4 min, using the GeneAmp PCR System 9700 (Applied Biosystems). Subsequently, direct sequencing was performed using 3730xl DNA Analyzer (Applied Biosystems). Sequence analyses were performed using Sequencher 5.2.0 software (Gene Codes, Ann Arbor, MI, USA).
Statistical analysis
Association studies were conducted using Fisher’s exact test for categorical variables and Mann–Whitney’s U test for continuous variables, and associations were considered statistically significant when the p-values were less than 0.05. If one of the cells in the contingency table was zero when calculating the odds ratio, to avoid error, the Haldane correction was applied by adding 0.5 to all the cells in the contingency table. Subsequently, univariate logistic regression analysis was carried out for the subanalysis by including only poor metabolizers (PMs), using an additive model of UGT1A1*6 to UGT1A1*28. For this, we assigned 0 for UGT1A1*28/*28, 1 for UGT1A1*6/*28, and 2 for UGT1A1*6/*6. Where correlation analysis was performed, Spearman r values were provided. Multivariate logistic regression analysis with a stepwise selection of variables was conducted to identify independent factors associated with ADRs due to irinotecan. In addition, we evaluated the diagnostic performance of the predictive model using the carrier status of UGT1A1*6/*6 and concomitant use of platinum-based antineoplastic drugs between cases and tolerant controls. Sensitivity, specificity, positive predictive value, negative predictive value, and number needed to screen (NNS) were calculated. Finally, a receiver-operating characteristic (ROC) curve was plotted with true positive rate (sensitivity) versus false positive rate (1-specificity), and area under curve (AUC) was used to evaluate how well the prediction model could distinguish between the risks of ADRs by irinotecan. The ROC curves of the prediction models were compared using DeLong’s test [20, 21]. All analyses were carried out using the R statistical environment 3.5.0 [22].
Results
A flow chart of patients included in this study is shown in Supplemental Fig. 1. Among 651 patients treated with irinotecan in the BioBank Japan, 15.7% developed grade 3–5 ADRs. The patient demographic details are summarized in Table 1. The proportions of cases and controls for each concomitant drug and drug classifications are shown. The details of the drug classifications (how each drug was grouped) are shown in Supplemental Table 1. The difference in percentages of cases and tolerant controls were statistically significant for lung cancer diagnoses, with cisplatin, vinorelbine, and platinum/others as concomitant drug names, and platinum-based antineoplastic and antimetabolites as concomitant drug groups (Table 1). The associations of the UGT1A1 diplotype with irinotecan-related ADRs are shown in Table 2. UGT1A1*6/*6 was the only diplotype with a statistically significant difference between cases and tolerant controls. UGT1A1*6/*28 and UGT1A1*28/*28 did not show significant associations with ADRs. The subanalysis comprised of only the PMs revealed a trend towards an increased risk of ADRs in patients with UGT1A1*6 (p-value 0.0092, odds ratio 4.39, 95% confidence interval 1.57–14.9). The analysis only among patients without lung cancer also showed that UGT1A1*6/*6 was the only diplotype with a statistically significant difference between cases and tolerant controls (Table S2). The stratified analyses were performed separately by platinum-based antineoplastics and antimetabolites, which also showed that UGT1A1*6/*6 was the only diplotype with a statistically significant difference (Tables S3 and S4). A summary describing the patients’ ADR grade and carrier status of the number of UGT1A1*6 is shown in Table S5. There are no correlations between UGT1A1*6 and ADR grade from 1 to 5 (r = 0.032). As shown in Table 3, PMs showed statistically significant associations with ADRs.
Since the proportion of patients with lung cancer or UGT1A1*6/*6 taking platinum-based antineoplastic drugs or antimetabolites is significantly different between cases and tolerant controls, we conducted a multiple logistic regression analysis using those variables (Table 4). In the constructed model incorporating the variables of lung cancer, UGT1A1*6/*6, platinum-based antineoplastic drugs, and antimetabolites predictors, the p-values were 0.27, 0.00036, 0.051, and 0.37, respectively. In a subsequent stepwise model, concomitant use of platinum-based antineoplastic drugs and UGT1A1*6/*6 remained significant. The predictive model can be expressed as below:
We drew the ROC curve for the prediction of ADR risk. The AUC was improved from 0.581 to 0.604 by adding UGT1A1*6/*6 to the concomitant use of platinum-based antineoplastic drugs, although it was not found to be statistically significant (p = 0.068) (Fig. 1).
Receiver-operating characteristic (ROC) curve to predict the risk of adverse drug reactions by irinotecan. The black line shows the ROC curve for the model using data from patients administered platinum-based antineoplastic drugs in addition to irinotecan. The area under the curve (AUC) of the black line equals 0.581 (95% confidence interval 0.529–0.633). The blue line shows the ROC curve for the UGT1A1*6/*6-based model. The AUC of the blue line equals 0.533 (95% confidence interval 0.506–0.560). The red line shows the ROC curve for the combined model of UGT1A1*6/*6 and patient use of platinum-based antineoplastic drugs. The AUC of the red line was 0.604 (95% confidence interval 0.550–0.659)
The diagnostic performance of the predictive model is shown in Table 5. At least 34.8% of the frequency of irinotecan-related ADRs was reported by Shiozawa et al. [23]. They reported the frequencies of major grade 3–4 ADRs such as leukopenia (34.8%), thrombocytopenia (12.4%), and diarrhea (10.1%) [23]. Their study was based on case report forms of 13,935 patients (94.1% of 14,802 patients registered) treated with irinotecan-based chemotherapy.
Discussion
In this study, we showed that UGT1A1*6/*6 is significantly associated with irinotecan-induced toxicities in the Japanese population, using one of the largest genetic databases, the Biobank Japan. Our key finding was that UGT1A1*6, rather than UGT1A1*28, contributes to ADR occurrence. Thus, we emphasized the importance of checking the carrier status of UGT1A1*6 in patients of Japanese backgrounds even outside of Japan.
Since the first report by Ando et al., a number of studies have examined the associations of pharmacokinetic parameters or clinical responses after administering irinotecan [24]. The associations between irinotecan-induced toxicities and UGT1A1 genetic polymorphisms are already widely known, and can be correlated with the therapeutic recommendations on drug labels by the Pharmaceuticals and Medical Devices Agency in Japan, the U.S. Food and Drug Administration, and Health Canada Santé Canada, and recommendations by the Dutch Pharmacogenetics Working Group and the French National Network of Pharmacogenetics [1, 2, 25,26,27]. However, they comment only or mainly on UG1A1*28 for irinotecan use because of the higher frequency of this allele in their own major ethnicities. Although UGT1A1 testing has been covered by the Japanese national health insurance for over 10 years, our recent survey revealed that UGT1A1 testing is rarely covered by major health insurers in the US because of insufficient evidence based on their medical policies [3]. Hence, using one of the largest databases, the BioBank Japan, we successfully showed significant association of UGT1A1*6/*6 with irinotecan-related ADRs, which could be meaningful because it would definitely add stronger evidence to support the positive associations.
UGT1A1 alleles with decreased activity could be associated with higher plasma concentrations of SN-38 and risks of ADRs. A number of studies in the Japanese population have shown associations with plasma concentrations since Araki et al. reported in 2006 that UGT1A1*6 was associated with higher ratios of the area under a plasma concentration-time curve (AUCSN-38/AUCSN-38 glucuronide) [28], which has been confirmed by other Japanese study groups [29,30,31]. Our findings corroborated the previously reported positive toxicity findings by Minami et al., specifically the positive associations of the UGT1A1*6 allele with neutropenia in Japanese patients, which has since been confirmed by multiple Japanese group studies [17, 30, 32,33,34,35,36,37,38,39,40]. However, these findings did not lead to a consensus on the utility of assessing UGT1A1*6 carrier status, thus resulting in no change to the official recommendations for the testing, as mentioned above. This is likely because of the small sample sizes included in each study, and the small number of previous studies by Japanese research groups that were likely included in previous systematic reviews and meta-analyses because of notable heterogeneities across the studies [4, 5, 7, 9,10,11].
UGT1A1*28 was the only allele that showed a positive association with irinotecan-induced toxicities in the Japanese population after the initial reports on the association of the UGT1A1 gene by Ando et al. [24]. Multiple subsequent studies have reported on the association of UGT1A1*6 [15, 24, 41]. Our findings showed no significant associations with UGT1A1*28; however, no case patients in our study had UGT1A1*28/*28. The frequency of UGT1A1*28 in the Japanese population was lower than that in Caucasians (0.051–0.135 and 0.271–0.380, respectively) [42,43,44], and the frequency of UGT1A1*28/*28 was quite low in the Japanese population (0.023); [45] hence, it was not surprising that our results could be related to small sample sizes even on using the data from the BioBank Japan.
Our results suggested that UGT1A1*6 contributes to decreased UGT1A1 activity and ADR occurrence more than UGT1A1*28 based on the fact that the subanalysis comprising only the PMs showed that UGT1A1*6 additively influenced the odds ratios. Multiple previous studies that investigated the associations of pharmacokinetic parameters could support our findings. For example, Minami et al. showed the medians of AUCSN-38 glucuronide/AUCSN-38 for UGT1A1 diplotypes that were 1.19 for UGT1A1*6/*6, 2.03 for *6/*28, and 3.65 for *28/*28 [17]. Satoh et al. [30] reported that the associations of UGT1A1 genotypes with AUCSN-38 glucuronide/AUCSN-38 were 1.21 ± 0.36 for UGT1A1*6/*6, 2.34 ± 0.82 for *6/*28, and 3.10 ± 1.82 for *28/*28. Therefore, it is possible that UGT1A1*6 decreases UGT1A1 activity more than UGT1A1*28, which is supported by previous meta-analyses by Chen et al. [9], wherein ADRs on irinotecan treatment were found to be more significantly associated with UGT1A1*6 than with UGT1A1*28. This would also support the current situation, where UGT1A1 testing is covered by the national health insurance in Japan because the UGT1A1*6 frequency is higher in Asian populations, who are hence more likely to experience more ADRs after using irinotecan than in other ethnicities.
Multiple logistic regression analyses showed that the concurrent use of platinum-based antineoplastic drugs was significantly associated with ADRs in addition to UGT1A1*6/*6. The platinum-based antineoplastic drugs could cause side effects such as cytopenia and diarrhea [46]. In addition, we corroborated the findings of the few studies on the Japanese population that have shown higher ADR incidence from the use of irinotecan plus platinum-based chemotherapy [47, 48]. Most importantly, the coefficient of UGT1A1*6/*6 was 1.868 (versus 0.6932 for platinum-based antineoplastic drugs), implying that UGT1A1*6/*6 could be the stronger variable in the multiple logistic regression analysis.
The diagnostic performance of the concurrent use of two risk factors, UGT1A1*6/*6 and platinum-based antineoplastic drugs, for ADRs was significant because the NNS was as low as 5.8. The area under the curve was improved to 0.604 by combining the two risk factors. Thus, we have identified a component that could explain ADRs by irinotecan; however, we must also consider other components that may be associated with ADRs. Considering these risk factors could help reduce the number of patients who suffer from the ADRs, thus achieving the purpose of preemptive pharmacogenomics testing performed to avoid ADRs.
Our study has some limitations. First, the dosages of irinotecan were not available for each patient in the BioBank Japan. However, we think that this limitation could be overcome by knowing their diagnoses and the names of the concomitantly used drugs, and assuming the regimens, which might not affect the conclusions in our study because our aim was to determine the overall associations by combining all patients who took irinotecan. Second, the information about whether or not the patients were tested for UGT1A1 before starting chemotherapy is not available. If patients carried UGT1A1*6 and/or *28 alleles, they might have received the lower starting doses of irinotecan. Thus, there is a potential of showing a higher percentage of tolerant controls for patients carrying UGT1A1*6 and/or *28 alleles, leading to a possible underestimation of the predictive values of the risk alleles in our study. Third, in our study cases, we did not observe UGT1A1*28/28, suggesting that the sample sizes must be increased; however, we believe that our study is the second largest study examining the associations of irinotecan-induced ADRs in the Japanese population.
In conclusion, we showed that the UGT1A1*6/*6 is significantly associated with ADRs due to irinotecan, and that UGT1A1*6 rather than UGT1A1*28 contributed to ADR occurrence. Knowing the UGT1A1*6 carrier status would be useful to predict the occurrence of ADRs with irinotecan use in Japan and around the world.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Pharmaceuticals and Medical Devices Agency—Drug label. 2019. https://www.pmda.go.jp/PmdaSearch/iyakuDetail/ResultDataSetPDF/800015_4240404A1040_1_11.
Food and Drug Administration—Drug label. 2014. https://www.accessdata.fda.gov/drugsatfda_docs/label/2014/020571s048lbl.pdf.
Hikino K, Fukunaga K, Mushiroda T. Gap between the US and Japan in coverage of pharmacogenomic biomarkers by health insurance programs: more coverage is needed in Japan. Drug Metab Pharmacokinet. 2018;33:243–9.
Zhang X, Yin JF, Zhang J, Kong SJ, Zhang HY, Chen XM. UGT1A1*6 polymorphisms are correlated with irinotecan-induced neutropenia: a systematic review and meta-analysis. Cancer Chemother Pharmacol. 2017;80:135–49.
Chen YJ, Hu F, Li CY, Fang JM, Chu L, Zhang X, et al. The association of UGT1A1*6 and UGT1A1*28 with irinotecan-induced neutropenia in Asians: a meta-analysis. Biomarkers. 2014;19:56–62.
Cheng L, Li M, Hu J, Ren W, Xie L, Sun ZP, et al. UGT1A1*6 polymorphisms are correlated with irinotecan-induced toxicity: a system review and meta-analysis in Asians. Cancer Chemother Pharmacol. 2014;73:551–60.
Yang Y, Zhou M, Hu M, Cui Y, Zhong Q, Liang L, et al. UGT1A1*6 and UGT1A1*28 polymorphisms are correlated with irinotecan-induced toxicity: a meta-analysis. Asia Pac J Clin Oncol. 2018;14:e479–e89.
Liu XH, Lu J, Duan W, Dai ZM, Wang M, Lin S, et al. Predictive value of UGT1A1*28 polymorphism In irinotecan-based chemotherapy. J Cancer. 2017;8:691–703.
Chen X, Liu L, Guo Z, Liang W, He J, Huang L, et al. UGT1A1 polymorphisms with irinotecan-induced toxicities and treatment outcome in Asians with Lung Cancer: a meta-analysis. Cancer Chemother Pharmacol. 2017;79:1109–17.
Campbell JM, Stephenson MD, Bateman E, Peters MD, Keefe DM, Bowen JM. Irinotecan-induced toxicity pharmacogenetics: an umbrella review of systematic reviews and meta-analyses. Pharmacogenomics J. 2017;17:21–8.
Han FF, Guo CL, Yu D, Zhu J, Gong LL, Li GR, et al. Associations between UGT1A1*6 or UGT1A1*6/*28 polymorphisms and irinotecan-induced neutropenia in Asian cancer patients. Cancer Chemother Pharmacol. 2014;73:779–88.
Hirata M, Kamatani Y, Nagai A, Kiyohara Y, Ninomiya T, Tamakoshi A, et al. Cross-sectional analysis of BioBank Japan clinical data: a large cohort of 200,000 patients with 47 common diseases. J Epidemiol. 2017;27:S9–S21.
Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, et al. Overview of the BioBank Japan Project: study design and profile. J Epidemiol. 2017;27:S2–S8.
Institute NC. Common toxicity criteria (CTC) Version 2.0 June 1, 1999. https://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcmanual_v4_10-4-99.pdf.
Ando Y, Saka H, Ando M, Sawa T, Muro K, Ueoka H, et al. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 2000;60:6921–6.
Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382–8.
Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497–504.
Sai K, Saeki M, Saito Y, Ozawa S, Katori N, Jinno H, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75:501–15.
Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high-throughput SNP typing system for genome-wide association studies. J Hum Genet. 2001;46:471–7.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. https://www.r-project.org/.
Shiozawa T, Tadokoro J, Fujiki T, Fujino K, Kakihata K, Masatani S, et al. Risk factors for severe adverse effects and treatment-related deaths in Japanese patients treated with irinotecan-based chemotherapy: a postmarketing survey. Jpn J Clin Oncol. 2013;43:483–91.
Ando Y, Saka H, Asai G, Sugiura S, Shimokata K, Kamataki T. UGT1A1 genotypes and glucuronidation of SN-38, the active metabolite of irinotecan. Ann Oncol. 1998;9:845–7.
Health Canada/Santé Canada. Product Monograph Camptosar. 2014. https://api.pharmgkb.org/v1/download/file/attachment/Irinotecan_HCSC_06_02_15.pdf.
Quaranta S, Thomas F. Pharmacogenetics of anti-cancer drugs: state of the art and implementation—recommendations of the French National Network of Pharmacogenetics. Therapie. 2017;72:205–15.
Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 2011;89:662–73.
Araki K, Fujita K, Ando Y, Nagashima F, Yamamoto W, Endo H, et al. Pharmacogenetic impact of polymorphisms in the coding region of the UGT1A1 gene on SN-38 glucuronidation in Japanese patients with cancer. Cancer Sci. 2006;97:1255–9.
Fujita K, Ando Y, Nagashima F, Yamamoto W, Eodo H, Araki K, et al. Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is associated with reduced activity for SN-38 in Japanese patients with cancer. Cancer Chemother Pharmacol. 2007;60:515–22.
Satoh T, Ura T, Yamada Y, Yamazaki K, Tsujinaka T, Munakata M, et al. Genotype-directed, dose-finding study of irinotecan in cancer patients with UGT1A1*28 and/or UGT1A1*6 polymorphisms. Cancer Sci. 2011;102:1868–73.
Hirose K, Yamashita K, Takada H, Kaneda N, Fukami K, Maruo E, et al. Usefulness of one-point plasma SN-38G/SN-38 concentration ratios as a substitute for UGT1A1 genetic information after irinotecan administration. Int J Clin Oncol. 2014;19:397–402.
Sai K, Saito Y, Sakamoto H, Shirao K, Kurose K, Saeki M, et al. Importance of UDP-glucuronosyltransferase 1A1*6 for irinotecan toxicities in Japanese cancer patients. Cancer Lett. 2008;261:165–71.
Onoue M, Terada T, Kobayashi M, Katsura T, Matsumoto S, Yanagihara K, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol. 2009;14:136–42.
Takano M, Kato M, Yoshikawa T, Sasaki N, Hirata J, Furuya K, et al. Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology. 2009;76:315–21.
Yamamoto N, Takahashi T, Kunikane H, Masuda N, Eguchi K, Shibuya M, et al. Phase I/II pharmacokinetic and pharmacogenomic study of UGT1A1 polymorphism in elderly patients with advanced non-small cell lung cancer treated with irinotecan. Clin Pharmacol Ther. 2009;85:149–54.
Nakamura Y, Soda H, Oka M, Kinoshita A, Fukuda M, Fukuda M, et al. Randomized phase II trial of irinotecan with paclitaxel or gemcitabine for non-small cell lung cancer: association of UGT1A1*6 and UGT1A1*27 with severe neutropenia. J Thorac Oncol. 2011;6:121–7.
Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K, et al. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol. 2011;41:477–82.
Hirasawa A, Zama T, Akahane T, Nomura H, Kataoka F, Saito K, et al. Polymorphisms in the UGT1A1 gene predict adverse effects of irinotecan in the treatment of gynecologic cancer in Japanese patients. J Hum Genet. 2013;58:794–8.
Hazama S, Mishima H, Tsunedomi R, Okuyama Y, Kato T, Takahashi K, et al. UGT1A1*6, 1A7*3, and 1A9*22 genotypes predict severe neutropenia in FOLFIRI-treated metastatic colorectal cancer in two prospective studies in Japan. Cancer Sci. 2013;104:1662–9.
Takahara N, Nakai Y, Isayama H, Sasaki T, Satoh Y, Takai D, et al. Uridine diphosphate glucuronosyl transferase 1 family polypeptide A1 gene (UGT1A1) polymorphisms are associated with toxicity and efficacy in irinotecan monotherapy for refractory pancreatic cancer. Cancer Chemother Pharmacol. 2013;71:85–92.
Ando Y, Ueoka H, Sugiyama T, Ichiki M, Shimokata K, Hasegawa Y. Polymorphisms of UDP-glucuronosyltransferase and pharmacokinetics of irinotecan. Ther Drug Monit. 2002;24:111–6.
Saeki M, Saito Y, Jinno H, Tohkin M, Kurose K, Kaniwa N, et al. Comprehensive UGT1A1 genotyping in a Japanese population by pyrosequencing. Clin Chem. 2003;49:1182–5.
Nishijima T, Tsuchiya K, Tanaka N, Joya A, Hamada Y, Mizushima D, et al. Single-nucleotide polymorphisms in the UDP-glucuronosyltransferase 1A-3′ untranslated region are associated with atazanavir-induced nephrolithiasis in patients with HIV-1 infection: a pharmacogenetic study. J Antimicrob Chemother. 2014;69:3320–8.
Premawardhena A, Fisher CA, Liu YT, Verma IC, de Silva S, Arambepola M, et al. The global distribution of length polymorphisms of the promoters of the glucuronosyltransferase 1 gene (UGT1A1): hematologic and evolutionary implications. Blood Cells Mol Dis. 2003;31:98–101.
Akiyama Y, Fujita K, Nagashima F, Yamamoto W, Endo H, Sunakawa Y, et al. Genetic testing for UGT1A1*28 and *6 in Japanese patients who receive irinotecan chemotherapy. Ann Oncol. 2008;19:2089–90.
Oun R, Moussa YE, Wheate NJ. The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton Trans. 2018;47:6645–53.
Nishikawa K, Murotani K, Fujitani K, Inagaki H, Akamaru Y, Tokunaga S, et al. A study of second-line irinotecan plus cisplatin vs. irinotecan alone in platinum-naive patients with early relapse of gastric cancer refractory to adjuvant S-1 monotherapy: exploratory subgroup analysis of the randomized phase III TRICS trial. Cancer Chemother Pharmacol. 2019;83:867–74.
Nishikawa K, Fujitani K, Inagaki H, Akamaru Y, Tokunaga S, Takagi M, et al. Randomised phase III trial of second-line irinotecan plus cisplatin versus irinotecan alone in patients with advanced gastric cancer refractory to S-1 monotherapy: TRICS trial. Eur J Cancer. 2015;51:808–16.
Acknowledgements
We appreciate all patients who participated in this study. The samples and data used for this study were provided from the BioBank Japan Project supported by the Japan Agency for Medical Research and Development (AMED) (Grant Number: JP19km0605001). This study was supported by grants from the Platform Program for Promotion of Genome Medicine funded by AMED (Grant Number: JP18km0405201). We express our gratitude to the staff of BioBank Japan for their assistance.
Author information
Authors and Affiliations
Contributions
KH, TO, MaK, and TM designed the study and analyzed and interpreted the data. KH wrote the paper. TO, CT, YK, YM, MiK, and TM performed the critical revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hikino, K., Ozeki, T., Koido, M. et al. Comparison of effects of UGT1A1*6 and UGT1A1*28 on irinotecan-induced adverse reactions in the Japanese population: analysis of the Biobank Japan Project. J Hum Genet 64, 1195–1202 (2019). https://doi.org/10.1038/s10038-019-0677-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0677-2
This article is cited by
-
Implementation of CYP2D6 copy-number imputation panel and frequency of key pharmacogenetic variants in Finnish individuals with a psychotic disorder
The Pharmacogenomics Journal (2022)
-
Genetic Analysis of UGT1A1 Polymorphisms Using Preserved Dried Umbilical Cord for Assessing the Potential of Neonatal Jaundice as a Risk Factor for Autism Spectrum Disorder in Children
Journal of Autism and Developmental Disorders (2022)
-
Higher Febuxostat Exposure Observed in Asian Compared with Caucasian Subjects Independent of Bodyweight
Clinical Pharmacokinetics (2021)
-
The Use of Subgroup Disproportionality Analyses to Explore the Sensitivity of a Global Database of Individual Case Safety Reports to Known Pharmacogenomic Risk Variants Common in Japan
Drug Safety (2021)