Key Points
-
Methods for purifying virus-like particles (VLPs) from microbial communities, procedures for amplifying the small quantities of DNA that are recovered from VLPs, advances in next-generation sequencing, and a number of new computational approaches have laid the foundations for a 'new age of phage', in which rapid progress is being made in characterizing the viral diversity and virus–bacterial host dynamics in the microbial communities residing in a broad range of habitats, including those associated with our human bodies.
-
Phage genomes have limited sequence conservation, making comparative genomics difficult. There is no conserved phylogenetic marker. Despite these obstacles, new techniques have been developed to classify phages.
-
The phage community in the human gut is complex but appears to be much more stable than those in other habitats, such as the ocean. Patterns of temporal and functional variation are being defined using metagenomics. Gnotobiotic animal models hold promise for further characterization of the role of phages in shaping the properties of the human gut microbiota, including its responses to various perturbations.
-
New insights about the microbial ecology in humans have been gleaned from comparative metagenomics studies and have rekindled an interest in phage therapy. Therapeutic goals may include enhancing the ability of probiotic consortia to establish themselves, and the addition of novel functions to the gut microbiome. Representative preclinical models are needed for proof-of-principle, proof-of-efficacy, dosing and safety tests.
Abstract
Over the past decade, researchers have begun to characterize viral diversity using metagenomic methods. These studies have shown that viruses, the majority of which infect bacteria, are probably the most genetically diverse components of the biosphere. Here, we briefly review the incipient rise of a phage biology renaissance, which has been catalysed by advances in next-generation sequencing. We explore how work characterizing phage diversity and lifestyles in the human gut is changing our view of ourselves as supra-organisms. Finally, we discuss how a renewed appreciation of phage dynamics may yield new applications for phage therapies designed to manipulate the structure and functions of our gut microbiomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hershey, A. D. & Chase, M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36, 39–56 (1952).
Crick, F. H., Barnett, L., Brenner, S. & Watts-Tobin, R. J. General nature of the genetic code for proteins. Nature 192, 1227–1232 (1961).
Cairns, J., Stent, G. S. & Watson, J. D. Phage and the Origins of Molecular Biology (Cold Spring Harbor Laboratory Press, 1992).
Mokili, J. L., Rohwer, F. & Dutilh, B. E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2, 63–77 (2012).
Breitbart, M. & Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 (2005).
Fernandes, P. Antibacterial discovery and development—the failure of success? Nature Biotech. 24, 1497–1503 (2006).
d'Herelle, F. Sur un microbe invisible antagoniste des bacilles dysenteriques. C. R. Acad. Sci. Ser. D 165, 373–375 (1917).
Sulakvelidze, A., Alavidze, Z. & Morris, J. G. Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659 (2001).
Levin, B. R. & Bull, J. J. Population and evolutionary dynamics of phage therapy. Nature Rev. Microbiol. 2, 166–173 (2004).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Rev. Genet. 11, 181–190 (2010).
Horvath, P. & Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 (2010). An overview of CRISPR-mediated defence mechanisms against phage attack.
Virgin, H. W., Wherry, E. J. & Ahmed, R. Redefining chronic viral infection. Cell 138, 30–50 (2009). A discussion of the nuanced role of the immune system in chronic viral infections.
Delwart, E. Animal virus discovery: improving animal health, understanding zoonoses, and opportunities for vaccine development. Curr. Opin. Virol. 2, 1–9 (2012).
Haynes, M. & Rohwer, F. in Metagenomics of the Human Body (ed. Nelson, K. E.) 63–77 (Springer, 2011).
Fox, G. E. et al. The phylogeny of prokaryotes. Science 209, 457–463 (1980).
Lane, D. J. et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA 82, 6955–6959 (1985).
Hugenholtz, P., Goebel, B. M. & Pace, N. R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774 (1998).
Culley, A. I., Lang, A. S. & Suttle, C. A. High diversity of unknown picorna-like viruses in the sea. Nature 424, 1054–1057 (2003).
Breitbart, M., Miyake, J. H. & Rohwer, F. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236, 249–256 (2004).
Hambly, E. et al. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl Acad. Sci. USA 98, 11411–11416 (2001).
Casjens, S. R. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8, 451–458 (2005).
Reyes, A. et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338 (2010). The finding that the human faecal virome in healthy individuals is not highly shared between family members, and exhibits surprising stability over the course of 1 year.
Minot, S. et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625 (2011). A longitudinal study of the impact of controlled diet changes on the human gut virome.
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Thurber, R. V., Haynes, M., Breitbart, M., Wegley, L. & Rohwer, F. Laboratory procedures to generate viral metagenomes. Nature Protoc. 4, 470–483 (2009). A protocol for isolating VLPs for subsequent metagenomic characterization.
Willner, D. et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4547–4553 (2011).
Rohwer, F., Seguritan, V., Choi, D. H., Segall, A. M. & Azam, F. Production of shotgun libraries using random amplification. BioTechniques 31, 108–112 (2001).
Breitbart, M. et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185, 6220–6223 (2003).
Shan, T. et al. The fecal virome of pigs on a high-density farm. J. Virol. 85, 11697–11708 (2011).
Yozwiak, N. L. et al. Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl. Trop. Dis. 6, e1485 (2012).
Li, L. et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84, 6955–6965 (2010).
Ge, X. et al. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in china. J. Virol. 86, 4620–4630 (2012).
Hutchison, C. A., Smith, H. O., Pfannkoch, C. & Venter, J. C. Cell-free cloning using φ29 DNA polymerase. Proc. Natl Acad. Sci. USA 102, 17332 (2005).
Kim, K. H. et al. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74, 5975–5985 (2008).
Lasken, R. S. & Stockwell, T. B. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7, 19 (2007).
Kim, K. H. & Bae, J. W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77, 7663–7668 (2011).
Andrews-Pfannkoch, C., Fadrosh, D. W., Thorpe, J. & Williamson, S. J. Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages. Appl. Environ. Microbiol. 76, 5039–5045 (2010).
Fadrosh, D. W., Andrews-Pfannkoch, C. & Williamson, S. J. Separation of single-stranded DNA, double-stranded DNA and RNA from an environmental viral community using hydroxyapatite chromatography. J. Vis. Exp. 2011, e3146 (2011).
Marine, R. et al. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl. Environ. Microbiol. 77, 8071–8079 (2011).
Nakamura, S. et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS ONE 4, e4219 (2009).
Wommack, K. E., Bhavsar, J. & Ravel, J. Metagenomics: read length matters. Appl. Environ. Microbiol. 74, 1453–1463 (2008).
Bibby, K., Viau, E. & Peccia, J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 52, 386–392 (2011).
Ng, T. F. et al. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE 6, e20579 (2011).
Pasic, L. et al. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics 10, 570 (2009).
Vega Thurber, R. L. et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl Acad. Sci. USA 105, 18413–18418 (2008).
Dinsdale, E. A. et al. Functional metagenomic profiling of nine biomes. Nature 452, 629–632 (2008).
Yang, J. et al. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J. Clin. Microbiol. 49, 3463–3469 (2011).
Xu, B. et al. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS Pathog. 7, e1002369 (2011).
Coetzee, B. et al. Deep sequencing analysis of viruses infecting grapevines: virome of a vineyard. Virology 400, 157–163 (2010).
Minot, S., Grunberg, S., Wu, G. D., Lewis, J. D. & Bushman, F. D. Hypervariable loci in the human gut virome. Proc. Natl Acad. Sci. USA 109, 3962–3966 (2012). The finding that hypervariable loci in the virome are predicted to encode Ig superfamily and C-type lectin folds.
Rohwer, F. & Thurber, R. V. Viruses manipulate the marine environment. Nature 459, 207–212 (2009). An overview of the interactions between marine viruses and their hosts.
Leplae, R., Lima-Mendez, G. & Toussaint, A. ACLAME: A CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 38, D57–D61 (2010).
Overbeek, R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005).
Sharon, I. et al. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 5, 1178–1190 (2011).
Ghosh, T. S., Mohammed, M. H., Komanduri, D. & Mande, S. S. ProViDE: a software tool for accurate estimation of viral diversity in metagenomic samples. Bioinformation 6, 91–94 (2011).
Lorenzi, H. A. et al. TheViral MetaGenome Annotation Pipeline(VMGAP): an automated tool for the functional annotation of viral metagenomic shotgun sequencing data. Stand. Genomic Sci. 4, 418–429 (2011).
Meyer, F. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008).
Roux, S. et al. Metavir: a web server dedicated to virome analysis. Bioinformatics 27, 3074–3075 (2011).
Sun, S. et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 39, D546–D551 (2011).
Breitbart, M. et al. Genomic analysis of uncultured marine viral communities. Proc. Natl Acad. Sci. USA 99, 14250–14255 (2002).
Angly, F. et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6, 41 (2005).
Angly, F. E. et al. The marine viromes of four oceanic regions. PLoS Biol. 4, e368 (2006).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009).
Turnbaugh, P. J. et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl Acad. Sci. USA 107, 7503–7508 (2010).
Hansen, E. E. et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl Acad. Sci. USA 108 (Suppl. 1), 4599–4606 (2011).
Caporaso, J. G. et al. Moving pictures of the human microbiome. Genome Biol. 12, R50 (2011).
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Kim, M. S., Park, E. J., Roh, S. W. & Bae, J. W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070 (2011).
Krupovic, M. & Forterre, P. Microviridae goes temperate: microvirus-related proviruses reside in the genomes of Bacteroidetes. PLoS ONE 6, e19893 (2011).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Stern, A., Mick, E., Tirosh, I., Sagy, O. & Sorek, R. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 25 Jun 2012 (doi:10.1101/gr.138297.112)
Breitbart, M. et al. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159, 367–373 (2008). An article that describes the rapid assembly and unstable features of the gut virome following birth.
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Barondess, J. J. & Beckwith, J. A bacterial virulence determinant encoded by lysogenic coliphage λ. Nature 346, 871–874 (1990).
Plunkett, G. 3rd, Rose, D. J., Durfee, T. J. & Blattner, F. R. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181, 1767–1778 (1999).
Markine-Goriaynoff, N. et al. Glycosyltransferases encoded by viruses. J. Gen. Virol. 85, 2741–2754 (2004).
Liu, M. et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295, 2091–2094 (2002).
Fraser, J. S., Yu, Z., Maxwell, K. L. & Davidson, A. R. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359, 496–507 (2006).
Zhang, T. et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4, e3 (2006).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Muegge, B. D. et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011).
Cann, A. J., Fandrich, S. E. & Heaphy, S. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30, 151–156 (2005).
Donaldson, E. F. et al. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84, 13004–13018 (2010).
Blinkova, O. et al. Novel circular DNA viruses in stool samples of wild-living chimpanzees. J. Gen. Virol. 91, 74–86 (2010).
Ng, T. F. et al. Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J. Gen. Virol. 92, 1318–1323 (2011).
Phan, T. G. et al. The fecal viral flora of wild rodents. PLoS Pathog. 7, e1002218 (2011).
van den Brand, J. M. et al. Metagenomic analysis of the viral flora of pine marten and European badger feces. J. Virol. 86, 2360–2365 (2012).
Dhillon, T. S., Dhillon, E. K., Chau, H. C., Li, W. K. & Tsang, A. H. Studies on bacteriophage distribution: virulent and temperate bacteriophage content of mammalian feces. Appl. Environ. Microbiol. 32, 68–74 (1976).
Berg Miller, M. E. et al. Phage–bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ. Microbiol. 14, 207–227 (2012).
Maura, D. et al. Intestinal colonization by enteroaggregative Escherichia coli supports long-term bacteriophage replication in mice. Environ. Microbiol. 28 Nov 2011 (doi:10.1111/j.1462-2920.2011.02644.x).
Fischetti, V. A., Nelson, D. & Schuch, R. Reinventing phage therapy: are the parts greater than the sum? Nature Biotech. 24, 1508–1511 (2006).
Lu, T. K. & Koeris, M. S. The next generation of bacteriophage therapy. Curr. Opin. Microbiol. 14, 524–531 (2011).
van Helvoort, T. The controversy between John H. Northrop and Max Delbrück on the formation of bacteriophage: bacterial synthesis or autonomous multiplication? Ann. Sci. 49, 545–575 (1992).
Calendar, R. L. The Bacteriophages (Oxford Univ. Press, 2005).
Kumarasamy, K. K. et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602 (2010).
Piddock, L. J. The crisis of no new antibiotics—what is the way forward? Lancet Infect. Dis. 12, 249–253 (2012).
Clokie, M. R. J. & Kropinski, A. M. Bacteriophages: Methods and Protocols (Humana Press, 2009).
Hehemann, J. H. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010).
Geier, M. R., Trigg, M. E. & Merril, C. R. Fate of bacteriophage lambda in non-immune germ-free mice. Nature 246, 221–223 (1973).
Schubbert, R., Renz, D., Schmitz, B. & Doerfler, W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc. Natl Acad. Sci. USA 94, 961–966 (1997).
Schubbert, R., Hohlweg, U., Renz, D. & Doerfler, W. On the fate of orally ingested foreign DNA in mice: chromosomal association and placental transmission to the fetus. Mol. Gen. Genet. 259, 569–576 (1998).
Geier, M. R. & Merril, C. R. Lambda phage transcription in human fibroblasts. Virology 47, 638–643 (1972).
Barry, M. A., Dower, W. J. & Johnston, S. A. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nature Med. 2, 299–305 (1996).
Dunn, I. S. Mammalian cell binding and transfection mediated by surface-modified bacteriophage lambda. Biochimie 78, 856–861 (1996).
Silverman, M. S., Davis, I. & Pillai, D. R. Success of self-administered home fecal transplantation for chronic Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 8, 471–473 (2010).
Chibani-Chennoufi, S. et al. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother. 48, 2558–2569 (2004).
Weiss, M. et al. In vivo replication of T4 and T7 bacteriophages in germ-free mice colonized with Escherichia coli. Virology 393, 16–23 (2009).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14 (2009).
Goodman, A. L. et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl Acad. Sci. USA 108, 6252–6257 (2011).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009).
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998).
Bergh, O., Borsheim, K. Y., Bratbak, G. & Heldal, M. High abundance of viruses found in aquatic environments. Nature 340, 467–468 (1989).
Clokie, M. R., Millard, A. D., Letarov, A. V. & Heaphy, S. Phages in nature. Bacteriophage 1, 31–45 (2011).
Fuhrman, J. A. Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999).
Azam, F. et al. The ecological role of water column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263 (1983).
Marston, M. F. et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc. Natl Acad. Sci. USA 109, 4544–4549 (2012).
Mann, N. H., Cook, A., Millard, A., Bailey, S. & Clokie, M. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424, 741 (2003).
Sullivan, M. B. et al. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4, e234 (2006).
Lindell, D., Jaffe, J. D., Johnson, Z. I., Church, G. M. & Chisholm, S. W. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438, 86–89 (2005).
Anderson, R. E., Brazelton, W. J. & Baross, J. A. Is the genetic landscape of the deep subsurface biosphere affected by viruses? Front. Microbiol. 2, 219 (2011).
Roossinck, M. J. The good viruses: viral mutualistic symbioses. Nature Rev. Microbiol. 9, 99–108 (2011). An excellent outline of beneficial virus–host interactions in a variety of species.
Roossinck, M. J. Changes in population dynamics in mutualistic versus pathogenic viruses. Viruses 3, 12–19 (2011).
Brown, S. P., Le Chat, L., De Paepe, M. & Taddei, F. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16, 2048–2052 (2006).
Brown, S. P., Inglis, R. F. & Tadddei, F. Evolutionary ecology of microbial wars: within-host competition and (incidental) virulence. Evol. Appl. 2, 32–39 (2009).
Moran, N. A., Degnan, P. H., Santos, S. R., Dunbar, H. E. & Ochman, H. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc. Natl Acad. Sci. USA 102, 16919–16926 (2005).
Oliver, K. M., Degnan, P. H., Hunter, M. S. & Moran, N. A. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994 (2009).
Xu, P. et al. Virus infection improves drought tolerance. New Phytol. 180, 911–921 (2008).
Marquez, L. M., Redman, R. S., Rodriguez, R. J. & Roossinck, M. J. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515 (2007).
Ophel, K. M., Bird, A. F. & Kerr, A. Association of bacteriophage particles with toxin production by Clavibacter toxicus, the causal agent of annual ryegrass toxicity. Phytopathology 83, 676–681 (1993).
Holtz, L. R., Finkbeiner, S. R., Kirkwood, C. D. & Wang, D. Identification of a novel picornavirus related to cosaviruses in a child with acute diarrhea. Virol. J. 5, 159 (2008).
Finkbeiner, S. R. et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 4, e1000011 (2008).
Finkbeiner, S. R. et al. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol. J. 6, 161 (2009).
Phan, T. G. et al. A third gyrovirus species in human feces. J. Gen. Virol. 93, 1356–1361 (2012).
Kapoor, A. et al. Multiple novel astrovirus species in human stool. J. Gen. Virol. 90, 2965–2972 (2009).
Kapoor, A. et al. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201, 1633–1643 (2010).
Victoria, J. G. et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 83, 4642–4651 (2009).
Rosario, K., Duffy, S. & Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch. Virol. 4 Jul 2012 (doi:10.1007/s00705-012-1391-y).
Rodriguez-Valera, F. et al. Explaining microbial population genomics through phage predation. Nature Rev. Microbiol. 7, 828–836 (2009). A discussion of the consequences of phage predation on microbial substrain diversity, presented as a constant-diversity dynamics model.
Lu, T. K. & Collins, J. J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl Acad. Sci. USA 106, 4629–4634 (2009).
Acknowledgements
Work from the authors' laboratories that is described in this Review was supported by the US National Institutes of Health (NIH) (grants DK78669, DK30292 and DK70977 to J.I.G. and grant GM095384 to F.L.R.) and by the Crohn's and Colitis Foundation of America. A.R. is the recipient of an International Fulbright Science and Technology Award. N.P.S. is a member of the Washington University Medical Scientist Training Program (MSTP), which is funded by NIH grant GM007200. Owing to space limitations, the authors were not able to cite many wonderful studies that are relevant to the topics covered.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- CRISPRs
-
(Clustered regularly interspaced short palindromic repeats). Widespread genetic systems in bacteria and archaea, consisting of multiple copies of palindromic repeats flanking short spacers of viral or plasmid origin. CRISPR elements provide acquired resistance to foreign DNA.
- Virus-like particles
-
(VLPs). Particles that can be recovered from microbial communities using physical separation methods such as density gradient ultracentrifugation and/or filtration. Purified VLPs have physical characteristics that resemble those of viruses, although their capacity for infection has to be subsequently defined.
- Virotype
-
A taxonomic classification that is typically based on a selected percentage identity threshold among viral reads, rather than on phylogenetic markers.
- Multiple displacement amplification
-
A method for exponential isothermal amplification of a DNA template using φ29 DNA polymerase and random primers. Exponential amplification is achieved by attachment of the polymerase to newly elongated fragments, coupled with the strong displacement activity of the enzyme on extension.
- Deep biosphere
-
The deepest oceanic regions in which life is supported.
- Prophages
-
Temperate phages in a host-incorporated state.
- α-diversity
-
Diversity, whether defined using taxonomic or functional characteristics, within a particular locale (habitat) at a particular moment in time.
- β-diversity
-
Diversity measured between samples or locales at a particular moment in time or over time.
- γ-diversity
-
A combination of alpha and beta diversity.
- Pan-genome
-
The global gene repertoire of a microbial species; defined by sequencing the genomes of isolates of that species obtained from a single or multiple habitats.
- Bacterial phylotypes
-
Taxonomic classifications that are based on phylogenetic markers, classically the 16S rRNA gene. Isolates can be arbitrarily assigned to a species-level phylotype if they share ≥97% sequence identity among their 16S rRNA genes.
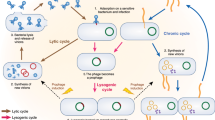
- Lysogenic
-
Pertaining to a temperate phage: a state in which linear (1:1) replication is achieved through integration of the phage genome into the chromosome of the bacterial host (or, more rarely, the phage exists as a plasmid within the host cell). The integrated phage transcribes genes that repress lytic action, and in some cases expresses genes that promote the fitness of the bacterial host.
- Kill-the-winner virus–bacterial host dynamic
-
A model for the population dynamics of phage–bacterium interactions; this model postulates that an increase in a host population (the winner) is followed by an increase in its corresponding phage predator, resulting in an increase in the rate at which the winner is killed.
- Coliphages
-
Phages that infect coliform bacteria, in particular Escherichia coli.
Rights and permissions
About this article
Cite this article
Reyes, A., Semenkovich, N., Whiteson, K. et al. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10, 607–617 (2012). https://doi.org/10.1038/nrmicro2853
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2853
This article is cited by
-
Eco-evolutionary dynamics of gut phageome in wild gibbons (Hoolock tianxing) with seasonal diet variations
Nature Communications (2024)
-
Emerging applications of phage therapy and fecal virome transplantation for treatment of Clostridioides difficile infection: challenges and perspectives
Gut Pathogens (2023)
-
Dietary D-xylose promotes intestinal health by inducing phage production in Escherichia coli
npj Biofilms and Microbiomes (2023)
-
Characterization of Phietavirus Henu 2 in the virome of individuals with acute gastroenteritis
Virus Genes (2023)
-
BoGH13ASus from Bacteroides ovatus represents a novel α-amylase used for Bacteroides starch breakdown in the human gut
Cellular and Molecular Life Sciences (2023)