Key Points
-
The movement of humans and commerce facilitate the emergence of new infectious diseases. Local pathogen populations can take advantage of human activities to create pandemics.
-
Recently emerged pathogens are very homogeneous owing to their recent derivation from another species or group. Even among highly similar subtypes, just a subset of strains becomes globally distributed owing to their high fitness.
-
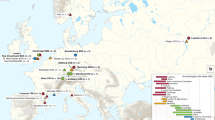
Bacillus anthracis (which causes anthrax) spread globally owing to spore contamination of animal products such as hair, hides and bones. Long-range transmission of anthrax does not occur naturally but rather depends on humans.
-
Yersinia pestis (which causes plague) evolved in Asia and was spread globally by human-mediated movement of infected rodents and their fleas. Regional and local ecological establishment is not uniform and is highly dependent on the make-up of local rodent populations.
-
Francisella tularensis (which causes tularaemia) has four subspecies, each with distinctive geographical distributions. Only F. tularensis subsp. holarctica is commonly found on multiple continents and represents a highly fit genotype that has clonally expanded in a circumpolar fashion.
-
Local pathogen populations may evolve 'hopeful monster' subtypes that can take advantage of ecological opportunities. In several cases, this opportunity has been human-mediated, long-distance dispersal with severe consequences for humans.
Abstract
The development of human civilizations and global commerce has led to the emergence and worldwide circulation of many infectious diseases. Anthrax, plague and tularaemia are three zoonotic diseases that have been intensely studied through genome characterization of the causative species and phylogeographical analyses. A few highly fit genotypes in each species represent the causative agents for most of the observed disease cases. Together, mutational and selective forces create highly adapted pathogens, but this must be coupled with ecological opportunities for global expansion. This Review describes the distributions of the bacteria that cause anthrax, plague and tularaemia and investigates the forces that created clonal structures in these species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pearson, T. et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl Acad. Sci. USA 101, 13536–13541 (2004).
Girard, J. M. et al. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proc. Natl Acad. Sci. USA 101, 8408–8413 (2004). This report characterizes plague outbreaks on a local and regional scale to reveal short temporal patterns in the transmission and evolution of the bacterium.
Vogler, A. J. et al. Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7. J. Bacteriol. 188, 4253–4263 (2006).
Price, L. B. et al. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob. Agents Chemother. 47, 2362–2365 (2003).
Vogler, A. J. et al. Molecular analysis of rifampin resistance in Bacillus anthracis and Bacillus cereus. Antimicrob. Agents Chemother. 46, 511–513 (2002).
Okinaka, R., Pearson, T. & Keim, P. Anthrax, but not Bacillus anthracis? PLoS Pathog. 2, e122 (2006).
Dragon, D. C. & Rennie, R. P. The ecology of anthrax spores: tough but not invincible. Can. Vet. J. 36, 295–301 (1995).
Pearson, T. et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl Acad. Sci. USA 101, 13536–13541 (2004). This article defines the main clade structure of B. anthracis as well as the nuances of phylogenetic reconstruction using whole-genome sequences.
Keim, P. et al. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179, 818–824 (1997).
Keim, P. et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182, 2928–2936 (2000).
Keim, P. et al. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4, 205–213 (2004).
Van Ert, M. N. et al. Global genetic population structure of Bacillus anthracis. PLoS ONE 2, e461 (2007). This is a comprehensive examination of B. anthracis global diversity based on molecular genetic analyses.
Simonson, T. S. et al. Bacillus anthracis in China and its relationship to worldwide lineages. BMC Microbiol. 9, 71 (2009).
Kenefic, L. J. et al. Pre-Columbian origins for North American anthrax. PLoS ONE 4, e4813 (2009).
Smith, K. L. et al. Bacillus anthracis diversity in Kruger National Park. J. Clin. Microbiol. 38, 3780–3784 (2000).
Fouet, A. et al. Diversity among French Bacillus anthracis isolates. J. Clin. Microbiol. 40, 4732–4734 (2002).
Gierczynski, R. et al. Intriguing diversity of Bacillus anthracis in eastern Poland – the molecular echoes of the past outbreaks. FEMS Microbiol. Lett. 239, 235–240 (2004).
Perry, R. D. & Fetherston, J. D. Yersinia pestis - etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66 (1997).
Gage, K. L. & Kosoy, M. Y. Natural history of plague: perspectives from more than a century of research. Annu. Rev. Entomol. 50, 505–528 (2005).
Achtman, M. et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA 96, 14043–14048 (1999).
Achtman, M. in Yersinia: Molecular and Cellular Biology (eds Carniel, E. & Hinnebusch, B. J.) 432 (Horizon Bioscience, Norwich, 2004).
Chain, P. S. G. et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA 101, 13826–13831 (2004).
Parkhill, J. et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413, 523–527 (2001).
Prentice, M. B. et al. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J. Bacteriol. 183, 2586–2594 (2001).
Achtman, M. et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl Acad. Sci. USA 101, 17837–17842 (2004). This paper defines the global population structure of Y. pestis based on multiple molecular genetic markers and, in particular, shows the phylogenetic plasticity of the traditional biochemical biovar designations.
Anisimov, A. P., Lindler, L. E. & Pier, G. B. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17, 434–464 (2004).
Zhou, D. et al. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 186, 5138–5146 (2004).
Drancourt, M. et al. Genotyping, Orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 10, 1585–1592 (2004).
Drancourt, M. et al. Yersinia pestis orientalis in remains of ancient plague patients. Emerg. Infect. Dis. 13, 332–333 (2007).
Deng, W. et al. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184, 4601–4611 (2002).
Auerbach, R. K. et al. Yersinia pestis evolution on a small timescale: comparison of whole genome sequences from North America. PLoS ONE 2, e770 (2007).
Hu, P. et al. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180, 5192–5202 (1998).
Dong, X., Ye, F. & Peng, H. Geographic distribution and feature of Yersinia pestis plasmid isolated from Yunnan province. Zhonghua Liu Xing Bing Xue Za Zhi 22, 344 (2001).
Filippov, A. A., Solodovnikov, N. S., Kookleva, L. M. & Protsenko, O. A. Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiol. Lett. 67, 45–48 (1990).
Galimand, M., Carniel, E. & Courvalin, P. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 50, 3233–3236 (2006).
Guiyoule, A. et al. Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 7, 43–48 (2001).
Keim, P., Johansson, A. & Wagner, D. M. Molecular epidemiology, evolution, and ecology of Francisella. Ann. NY Acad. Sci. 1105, 30–66 (2007). This paper provides a broad overview of F. tularensis ecology and evolution.
Farlow, J. et al. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39, 3186–3192 (2001).
Johansson, A. et al. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186, 5808–5818 (2004).
Farlow, J. et al. Francisella tularensis in the United States. Emerg. Infect. Dis. 11, 1835–1841 (2005).
Staples, J. E., Kubota, K. A., Chalcraft, L. G., Mead, P. S. & Petersen, J. M. Epidemiologic and molecular analysis of human tularemia, United States, 1964–2004. Emerg. Infect. Dis. 12, 1113–1118 (2006).
Gurycova, D. First isolation of Francisella tularensis subsp. tularensis in Europe. Eur. J. Epidemiol. 14, 797–802 (1998).
Chaudhuri, R. R. et al. Genome sequencing shows that European isolates of Francisella tularensis subspecies tularensis are almost identical to US laboratory strain Schu S4. PLoS ONE 2, e352 (2007).
Vogler, A. J. et al. Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J. Bacteriol. 191, 2474–2484 (2009). This paper defines the global population patterns of F. tularensis and, in particular, F. tularensis subsp. holarctica .
Belding, D. L. & Merrill, B. Tularemia in imported rabbits in Massachusetts. N. Engl. J. Med. 224, 1085–1087 (1941).
Barns, S. M., Grow, C. C., Okinaka, R. T., Keim, P. & Kuske, C. R. Detection of diverse new Francisella-like bacteria in environmental samples. Appl. Environ. Microbiol. 71, 5494–5500 (2005).
Mörner, T. & Addison, E. in Infectious Diseases of Wild Animals (eds Williams, E. S. & Barker, I. K.) 303–312 (Iowa State Univ. Press, Ames, 2001).
Abd, H., Johansson, T., Golovliov, I., Sandstrom, G. & Forsman, M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69, 600–606 (2003).
Berdal, B. P., Mehl, R., Meidell, N. K., Lorentzen-Styr, A. M. & Scheel, O. Field investigations of tularemia in Norway. FEMS Immunol. Med. Microbiol. 13, 191–195 (1996).
Zhou, D., Han, Y., Song, Y., Huang, P. & Yang, R. Comparative and evolutionary genomics of Yersinia pestis. Microbes Infect. 6, 1226–1234 (2004).
Nakazawa, Y. et al. Climate change effects on plague and tularemia in the United States. Vector Borne Zoonotic Dis. 7, 529–540 (2007).
Freedman, M. L. & Thorpe, M. E. Anthrax: a case report and a short review of anthrax in Australia. Med. J. Aust. 1, 154–157 (1969).
Blackburn, J. K., McNyset, K. M., Curtis, A. & Hugh-Jones, M. E. Modeling the geographic distribution of Bacillus anthracis, the causative agent of anthrax disease, for the contiguous United States using predictive ecologic niche modeling. Am. J. Trop. Med. Hyg. 77, 1103–1110 (2007).
Neerinckx, S., Peterson, A., Gulinck, H., Deckers, J. & Leirs, H. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int. J. Health Geogr. 7, 54 (2008).
Peterson, A. Biogeography of diseases: a framework for analysis. Naturwissenschaften 95, 483–491 (2008).
Goldschmidt, R. The Material Basis of Evolution (Yale Univ. Press, New Haven, 1940).
Gould, S. The return of hopeful monsters. Nat. Hist. 86, 22–30 (1977).
Okinaka, R. et al. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 87, 261–262 (1999).
Okinaka, R. T. et al. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181, 6509–6515 (1999).
Sebbane, F., Jarrett, C., Gardner, D., Long, D. & Hinnebusch, B. J. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77, 1222–1229 (2009).
Sodeinde, O. A. et al. A surface protease and the invasive character of plague. Science 258, 1004–1007 (1992).
Hinnebusch, B. J. et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296, 733–735 (2002).
Runco, L. M., Myrczek, S., Bliska, J. B. & Thanassi, D. G. Biogenesis of the fraction 1 capsule and analysis of the ultrastructure of Yersinia pestis. J. Bacteriol. 190, 3381–3385 (2008).
Cornelis, G. R. et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352 (1998).
Larsson, P. et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nature Genet. 37, 153–159 (2005).
Papagrigorakis, M. J., Yapijakis, C., Synodinos, P. N. & Baziotopoulou-Valavani, E. DNA examination of ancient dental pulp incriminates typhoid fever as a probable cause of the Plague of Athens. Int. J. Infect. Dis. 10, 206–214 (2006).
Drancourt, M. & Raoult, D. Molecular insights into the history of plague. Microbes Infect. 4, 105–109 (2002).
Wiechmann, I. & Grupe, G. Detection of Yersinia pestis DNA in two early medieval skeletal finds from Aschheim (Upper Bavaria, 6th century A.D.). Am. J. Phys. Anthropol. 126, 48–55 (2005).
Acknowledgements
We are grateful to J. Foster, R. Okinaka and T. Pearson for their comments. This work was supported by the US National Institutes of Health (GM060795, AI070183), the Pacific-Southwest Region Center of Excellence (AI065359) and the Department of Homeland Security Science and Technology Directorate (NBCH2070001 and HSHQDC-08-C00158).
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Mutation rate
-
The frequency with which nucleotide changes occur in a genome, usually standardized by events per generation.
- Horizontal gene transfer
-
The movement of DNA from the genome of one organism into that of another; also known as lateral gene transfer. The three primary mechanisms are transformation, which involves uptake of naked DNA from the environment by a bacterium, transduction, in which phages transfer DNA between organisms, and conjugation, which involves the transfer of DNA directly from bacterium to bacterium.
- Neutral genetic variation
-
Polymorphic genetic features that have no impact on the survival or reproductive success of the organism.
- Fixation
-
A genetic phenomenon in which a previously polymorphic feature takes on the same state in every individual in a population and, hence, is no longer polymorphic.
- Clonal organism
-
An organism that reproduces only by the division of somatic cells, without gene transfer or recombination. In this situation, genetic diversity is generated by mutation alone and not by gene transfer or recombination.
- Recently emerged pathogen
-
A pathogen species, subspecies, population or genotype with a short evolutionary history. It emerges from another species or population, possibly owing to a mutation that increases its fitness and leads to an increase in its frequency and distribution. The recent emergence of such pathogens is often inferred from the fact that there is little genetic diversity among individual isolates.
- Host shifting
-
Acquiring the capacity to infect a new host species and perhaps becoming adapted specifically to that new host.
- Phylogeography
-
Combining phylogenetics and geography to understand the geographical distribution of evolutionary patterns in a given organism.
- Phylogenetically informative characters
-
Polymorphic features (for example, a locus with multiple alleles) that have a genotype (for example, an allele) shared by two or more members (for example, isolates or strains) of the study group (for example, a population or a species), providing insights into the evolutionary relationships among members.
- Homoplasy
-
The occurrence of the same genotype in two organisms that is due not to shared descent but rather to other forces, such as a reverse mutation or recombination.
- Pleiotropic mutation
-
A single genetic change that leads to multiple phenotypes; for example, a regulatory mutation may affect many different genes and, therefore, the multiple phenotypes associated with those different genes.
- Obligate pathogen
-
An organism that can only survive in a host or vector and cannot persist directly in the environment.
- Sylvatic
-
Pertaining to wild animals as opposed to commensal animals, which are associated with humans.
- Phylogeny
-
A model of the evolutionary history of a set of individuals, species or other taxonomic units. Phylogenies are hypotheses that are based on the analysis of phylogenetically informative characters that arose as the organisms evolved. They are traditionally used to understand the evolution of species and other taxonomic units but are applicable to clonally propagating populations as well.
- Opportunistic pathogen
-
An organism that is normally benign but, given the right situation (for example, an immunocompromised host), can cause disease. Often humans or other hosts that are occasionally infected by these organisms are not important to the overall lifestyle of these pathogens and are dead-end hosts.
Rights and permissions
About this article
Cite this article
Keim, P., Wagner, D. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nat Rev Microbiol 7, 813–821 (2009). https://doi.org/10.1038/nrmicro2219
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2219
This article is cited by
-
First genome sequencing and comparative analyses of Corynebacterium pseudotuberculosis strains from Mexico
Standards in Genomic Sciences (2018)
-
Tularemia from a One Health Perspective
Current Clinical Microbiology Reports (2017)
-
Ecology of Tularemia in Central US Endemic Region
Current Tropical Medicine Reports (2016)
-
Increased knowledge of Francisella genus diversity highlights the benefits of optimised DNA-based assays
BMC Microbiology (2012)
-
A Revised Timeline for the Origin of Plasmodium falciparum as a Human Pathogen
Journal of Molecular Evolution (2011)