Abstract
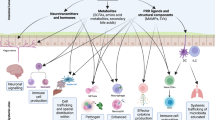
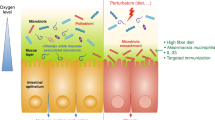
The mammalian intestine is home to a complex community of trillions of bacteria that are engaged in a dynamic interaction with the host immune system. Determining the principles that govern host–microbiota relationships is the focus of intense research. Here, we describe how the intestinal microbiota is able to influence the balance between pro-inflammatory and regulatory responses and shape the host's immune system. We suggest that improving our understanding of the intestinal microbiota has therapeutic implications, not only for intestinal immunopathologies but also for systemic immune diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009).
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006).
Eberl, G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 5 May 2010 (doi:10.1038/mi.2010.20).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Moran, N. A., McCutcheon, J. P. & Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190 (2008).
Garrett, W. S., Gordon, J. I. & Glimcher, L. H. Homeostasis and inflammation in the intestine. Cell 140, 859–870 (2010).
Hooper, L. V. & Macpherson, A. J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature Rev. Immunol. 10, 159–169 (2010).
Eberl, G. & Lochner, M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2, 478–485 (2009).
Rescigno, M. & Di Sabatino, A. Dendritic cells in intestinal homeostasis and disease. J. Clin. Invest. 119, 2441–2450 (2009).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009).
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L. & Hooper, L. V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl Acad. Sci. USA 105, 20858–20863 (2008).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).
Lotz, M. et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 203, 973–984 (2006).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Fagarasan, S. et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298, 1424–1427 (2002).
Malamut, G. et al. The enteropathy associated with common variable immunodeficiency: the delineated frontiers with celiac disease. Am. J. Gastroenterol. 15 Jun 2010 (doi:10.1038/ajg.2010.214).
Bouma, G. & Strober, W. The immunological and genetic basis of inflammatory bowel disease. Nature Rev. Immunol. 3, 521–533 (2003).
Glocker, E. O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Talham, G. L., Jiang, H. Q., Bos, N. A. & Cebra, J. J. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 67, 1992–2000 (1999).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Brandl, K. et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008).
Klaasen, H. L. et al. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab. Anim. 27, 141–150 (1993).
Lupp, C. et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129 (2007).
Stecher, B. et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Snel, J. et al. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int. J. Syst. Bacteriol. 45, 780–782 (1995).
Stepankova, R. et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211 (2007).
Wu, H.-S. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Lee, Y. K., Menezes, J. S., Umesaki, Y. & Mazmanian, S. K. Microbes and health sackler colloquium: proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 28 Jul 2010 (doi:10.1073/pnas.10000.82107).
Chow, J. & Mazmanian, S. K. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276 (2010).
Polk, D. B. & Peek, R. M. Jr. Helicobacter pylori: gastric cancer and beyond. Nature Rev. Cancer 10, 403–414 (2010).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Med. 15, 1016–1022 (2009).
Carvalho, F. A. et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206, 2179–2189 (2009).
Darfeuille-Michaud, A. et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127, 412–421 (2004).
Barnich, N. et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574 (2007).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Sokol, H. et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 12, 106–111 (2006).
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389 (2005).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Garrett, W. S. et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell 16, 208–219 (2009).
Garrett, W. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally-transmitted colitis. Cell Host Microbe (in the press).
Konrad, A., Cong, Y., Duck, W., Borlaza, R. & Elson, C. O. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology 130, 2050–2059 (2006).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009).
Bauer, H., Horowitz, R. E., Levenson, S. M. & Popper, H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 42, 471–483 (1963).
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O. & Kasper, D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Clarke, T. B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature Med. 16, 228–231 (2010).
Noverr, M. C. & Huffnagle, G. B. The 'microflora hypothesis' of allergic diseases. Clin. Exp. Allergy 35, 1511–1520 (2005).
Sjogren, Y. M., Jenmalm, M. C., Bottcher, M. F., Bjorksten, B. & Sverremark-Ekstrom, E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin. Exp. Allergy 39, 518–526 (2009).
Kuitunen, M. et al. Probiotics prevent IgE-associated allergy until age 5 years in cesarean-delivered children but not in the total cohort. J. Allergy Clin. Immunol. 123, 335–341 (2009).
Penders, J., Stobberingh, E. E., van den Brandt, P. A. & Thijs, C. The role of the intestinal microbiota in the development of atopic disorders. Allergy 62, 1223–1236 (2007).
Noverr, M. C., Falkowski, N. R., McDonald, R. A., McKenzie, A. N. & Huffnagle, G. B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect. Immun. 73, 30–38 (2005).
Bashir, M. E., Louie, S., Shi, H. N. & Nagler-Anderson, C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J. Immunol. 172, 6978–6987 (2004).
Vaahtovuo, J., Munukka, E., Korkeamaki, M., Luukkainen, R. & Toivanen, P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 35, 1500–1505 (2008).
Gray, D. H., Gavanescu, I., Benoist, C. & Mathis, D. Danger-free autoimmune disease in Aire-deficient mice. Proc. Natl Acad. Sci. USA 104, 18193–18198 (2007).
Hase, K. et al. Activation-induced cytidine deaminase deficiency causes organ-specific autoimmune disease. PLoS ONE 3, e3033 (2008).
Maldonado, M. A. et al. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. J. Immunol. 162, 6322–6330 (1999).
Sinkorova, Z., Capkova, J., Niederlova, J., Stepankova, R. & Sinkora, J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2k) male mice. Hum. Immunol. 69, 845–850 (2008).
Abdollahi-Roodsaz, S. et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 118, 205–216 (2008).
Breban, M. A., Moreau, M. C., Fournier, C., Ducluzeau, R. & Kahn, M. F. Influence of the bacterial flora on collagen-induced arthritis in susceptible and resistant strains of rats. Clin. Exp. Rheumatol. 11, 61–64 (1993).
Wen, L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 (2008).
Giraud, A. et al. Dissecting the genetic components of adaptation of Escherichia coli to the mouse gut. PLoS Genet. 4, e2 (2008).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008).
Backhed, F. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004).
Samuel, B. S. et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl Acad. Sci. USA 105, 16767–16772 (2008).
Turnbaugh, P. J. et al. The human microbiome project. Nature 449, 804–810 (2007).
Vijay-Kumar, M. et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231 (2010).
Peyrin-Biroulet, L. et al. Peroxisome proliferator-activated receptor γ activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl Acad. Sci. USA 107, 8772–8777 (2010).
Kumar, A. et al. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 26, 4457–4466 (2007).
Dubuquoy, L. et al. Impaired expression of peroxisome proliferator-activated receptor γ in ulcerative colitis. Gastroenterology 124, 1265–1276 (2003).
Kelly, D. et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nature Immunol. 5, 104–112 (2004).
Chieppa, M., Rescigno, M., Huang, A. Y. & Germain, R. N. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med. 203, 2841–2852 (2006).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Tsuji, M. et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science 323, 1488–1492 (2009).
Cerutti, A. & Rescigno, M. The biology of intestinal immunoglobulin A responses. Immunity 28, 740–750 (2008).
Coombes, J. L. & Powrie, F. Dendritic cells in intestinal immune regulation. Nature Rev. Immunol. 8, 435–446 (2008).
Atarashi, K. et al. ATP drives lamina propria TH17 cell differentiation. Nature 455, 808–812 (2008).
Zhou, L., Chong, M. M. & Littman, D. R. Plasticity of CD4+ T cell lineage differentiation. Immunity 30, 646–655 (2009).
Keilbaugh, S. A. et al. Activation of RegIIIβ/γ and interferon γ expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut 54, 623–629 (2005).
Umesaki, Y., Okada, Y., Matsumoto, S., Imaoka, A. & Setoyama, H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 39, 555–562 (1995).
Mayer, L. Evolving paradigms in the pathogenesis of IBD. J. Gastroenterol. 45, 9–16 (2010).
Acknowledgements
The authors thank W. Garrett for sharing unpublished data. Their work is supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut National de la Recherche Agronomique (INRA) and the Agence Nationale de la Recherche and Fondation Princesse Grace. The authors are partners of the European Community networks Cross-Talk (contract number PITN-GA-2008-215553) and Tornado (FP7 222720).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- Ankylosing enthesopathy
-
An inflammatory autoimmune disease of the joints that naturally occurs in mice on a C57BL/10 genetic background; the disease is similar to human ankylosing spondylitis. The pathology is characterized by the proliferation of cartilage and connective tissue, which culminates in ankylosis of the joints.
- Germinal centres
-
Highly specialized and dynamic microenvironments that are located in secondary lymphoid tissues and give rise to secondary B cell follicles during an immune response. Germinal centres are the main sites of B cell proliferation and differentiation, which leads to the generation of memory B cells and plasma cells that produce high-affinity antibodies.
- Gnotobiotic mice
-
Germ-free mice are born and raised in sterile isolators and are devoid of colonization by any microorganisms, but after they have been experimentally colonized by known bacteria, they are said to be gnotobiotic. They are kept in isolators to control their bacterial status.
- IgE-associated allergies
-
Type 1 hypersensitivity reactions that are mediated by IgE, which induces mast cell activation and degranulation. Such immune reactions are seen in asthma, allergic rhinitis, systemic anaphylaxis and food allergies.
- Obligate and facultative symbionts
-
Obligate microbial symbionts need to colonize a host to develop and multiply, unlike facultative microbial symbionts, which can also develop outside a host.
- Pathobionts
-
Microbial symbionts that can cause defined disease in predisposed hosts following changes in the gastrointestinal environment.
- Microbiome
-
The whole genome of all of the microorganisms that colonize a specific environment.
- Peyer's patches
-
Collections of lymphoid follicles that are located in the intestinal mucosa and are particularly abundant in the ileal mucosa. Together with mesenteric lymph nodes, they form the inductive compartment for intestinal immune responses.
- Proteobacteria
-
Gram-negative microorganisms that colonize very distinct environments and are the second largest group of bacteria on earth. Proteobacteria that colonize the intestine include commensal, pathogenic and opportunistic species, such as Salmonella, Shigella and Helicobacter spp. and Escherichia coli strains. In healthy adults, proteobacteria represent less than 1% of the enteric microbiota, but they are a major cause of intestinal and extraintestinal diseases.
- Type VI secretion system
-
(T6SS). Like T3SS and T4SS, T6SS is a multi-subunit complex that acts like a 'needle and syringe' to translocate bacterial products across the double-membrane of Gram-negative bacteria into the cytoplasm of eukaryotic cells.
- Xenobiotics
-
Chemical compounds that are foreign to a living organism and that can be toxic, even at low concentrations.
Rights and permissions
About this article
Cite this article
Cerf-Bensussan, N., Gaboriau-Routhiau, V. The immune system and the gut microbiota: friends or foes?. Nat Rev Immunol 10, 735–744 (2010). https://doi.org/10.1038/nri2850
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri2850
This article is cited by
-
Targeting the Gut Microbiome to Treat Cardiometabolic Disease
Current Atherosclerosis Reports (2024)
-
Gut microbiota in COVID-19: key microbial changes, potential mechanisms and clinical applications
Nature Reviews Gastroenterology & Hepatology (2023)
-
An antioxidant ameliorates allergic airway inflammation by inhibiting HDAC 1 via HIF-1α/VEGF axis suppression in mice
Scientific Reports (2023)
-
Captivity and Animal Microbiomes: Potential Roles of Microbiota for Influencing Animal Conservation
Microbial Ecology (2023)
-
High-fat diet alters intestinal microbiota and induces endoplasmic reticulum stress via the activation of apoptosis and inflammation in blunt snout bream
Fish Physiology and Biochemistry (2023)