Abstract
The chromosome 4 cluster of GABAA receptor genes is predominantly expressed in the brain reward circuitry and this chromosomal region has been implicated in linkage scans for alcoholism. Variation in one chromosome 4 gene, GABRA2, has been robustly associated with alcohol use disorders (AUD) although no functional locus has been identified. As HapMap data reveal moderate long-distance linkage disequilibrium across GABRA2 and the adjacent gene, GABRG1, it is possible that the functional locus is in GABRG1. We genotyped 24 SNPs across GABRG1 and GABRA2 in two population isolates: 547 Finnish Caucasian men (266 alcoholics) and 311 community-derived Plains Indian men and women (181 alcoholics). In both the Plains Indians and the Caucasians: (1) the GABRG1 haplotype block(s) did not extend to GABRA2; (2) GABRG1 haplotypes and SNPs were significantly associated with AUD; (3) there was no association between GABRA2 haplotypes and AUD; (4) there were several common (⩾0.05) haplotypes that spanned GABRG1 and GABRA2 (341 kb), three of which were present in both populations: one of these ancestral haplotypes was associated with AUD, the other two were more common in non-alcoholics; this association was determined by GABRG1; (5) in the Finns, three less common (<0.05) extended haplotypes showed an association with AUD that was determined by GABRA2. Our results suggest that there are likely to be independent, complex contributions from both GABRG1 and GABRA2 to alcoholism vulnerability.
Similar content being viewed by others
INTRODUCTION
A substantial body of evidence from preclinical studies has implicated GABAA receptors in the acute and chronic effects of ethanol including tolerance, dependence, and withdrawal (reviewed in Enoch, 2008; Krystal et al, 2006). Rapid synaptic inhibition is mediated through GABAA receptors that are ligand-gated, chloride ion channels formed by pentameric complexes composed of subunits (α, β, γ, δ, ɛ, π, and ρ), each of which have several isoforms (Barnard et al, 1998). Genes for the GABAA receptor subunit isoforms are clustered in several chromosomal regions. Expression of the various subunit isoforms varies across brain locations and during development. The mRNAs from the chromosome 4 cluster genes (GABRA2, GABRA4, GABRB1, and GABRG1) predominate in rat embryo but these genes are generally downregulated in the adult rat except in the hippocampus and in dopamine neurons in the substantia nigra and the ventral tegmental area where they are highly expressed (Okada et al, 2004; Steiger and Russek, 2004; Wisden et al, 1992). This suggests that the chromosome 4 gene cluster may be implicated in addiction. Indeed, earlier genome-wide scans in American Indians and Caucasians have provided evidence for linkage of alcohol dependence to the location of the chromosome 4 GABAA gene cluster (Long et al, 1998; Reich et al, 1998; Zinn-Justin and Abel, 1999). Subsequently, several studies have found haplotype and SNP associations between GABRA2 and alcoholism. All these studies, together with HapMap, have identified the same two GABRA2 haplotype blocks, at least within Caucasians, American Indians, and Asians. The significant association signals for alcoholism have been with two common yin-yang haplotypes within the haplotype block that extends downstream from intron 3 in the direction of the adjacent GABRG1 gene (Agrawal et al, 2006; Covault et al, 2004; Edenberg et al, 2004; Enoch et al, 2006a; Fehr et al, 2006; Lappalainen et al, 2005; Soyka et al, 2008). However, other studies have found no GABRA2 association with alcoholism (Covault et al, 2008, Project MATCH data set; Drgon et al, 2006; Matthews et al, 2007).
As yet, no functional GABRA2 locus has been identified. Moreover, a recent study has shown significant haplotype and SNP association with alcoholism in a haplotype block that extends from the intergenic region between GABRA2 and GABRG1 up to GABRG1 intron 3 in two large groups of US Caucasians but not in African Americans (Covault et al, 2008). Thus, it is possible that the apparent GABRA2 association with alcoholism may be due to linkage disequilibrium (LD) with a functional variant in GABRG1. The aim of our study was therefore first to examine the association between GABRG1 and alcoholism in Finnish Caucasians and Plains American Indians, two population isolates in which we had previously found a GABRA2 association with alcoholism that was mediated by anxiety (Enoch et al, 2006a). Second, as HapMap (http://www.hapmap.org/) has identified long-distance LD across the two adjacent genes, we investigated whether there were long-distance haplotypes spanning GABRA2 and GABRG1 that were common to both population isolates, ie ancestral haplotypes. To this end we genotyped 24 SNPs spanning GABRG1 from one intergenic region to the other and included two previously genotyped tag SNPs for the GABRA2 risk haplotypes.
PARTICIPANTS AND METHODS
Plains American Indians
Volunteers (311 total; 180 women, 131 men) were recruited from a Plains Indian tribe living in rural Oklahoma. The mean (SD) ages of participants were: women, 44.0 (14.9) years; men, 41.7 (12.9) years. The study was approved by the Plains Indian Tribal Council. Probands were initially ascertained at random from the tribal register and the families of alcoholic probands were extended. Exclusion criteria included a history of brain trauma and neurological diseases, together with current use of psychotropic medications and evidence of alcohol intoxication or withdrawal at the time of testing. The study was carefully explained to participants and written informed consent was obtained according to a human research protocol approved by the human research committee of the National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institutes of Health (NIH). The protocol and consent forms were also formally approved by the Plains Indian Tribal Council.
Blind-rated DSM-III-R lifetime psychiatric diagnoses (American Psychiatric Association, 1987) were derived from the Schedule for Affective Disorders and Schizophrenia—Lifetime version (SADS-L). A clinical social worker (BA), knowledgeable in the tribe's customs and culture, conducted the SADS-L interviews. The prevalence of lifetime alcohol use disorders (AUD, nearly all alcohol dependence) in this sample was 73% in men and 47% in women (Enoch et al, 2006b) however 63% of the alcoholics had been abstinent for at least 1 year. The sample included 181 alcoholics (97 men and 84 women) and 130 non-alcoholics (34 men and 96 women).
Finnish Caucasians
Finnish Caucasians are regarded as a population isolate. The sample from Helsinki, Finland, has been described in detail elsewhere (Lappalainen et al, 1998). In total, 547 men were genotyped: 266 alcoholics and 281 non-alcoholics. The sample was comprised of 173 incarcerated alcoholic criminal offenders, 159 relatives (93 alcoholic, 66 non-alcoholic), and 215 population controls. Of the alcoholics, 217 had alcohol dependence and 49 had alcohol abuse. Mean ages were: alcoholics, 34.4 years (SD=10.4); non-alcoholics: 33.1 years (SD=12.0).
Written informed consent was obtained according to human research protocols approved by the human research committees of NIAAA and the National Institute of Mental Health, NIH, the Department of Psychiatry, University of Helsinki, and the University of Helsinki Central Hospital, Helsinki, Finland.
The Structured Clinical Interview for DSM-III-R was administered by psychiatrists to both alcoholics and controls and blind-rated DSM-III-R psychiatric diagnoses were obtained. Individuals with major psychotic episodes were excluded.
Genotyping
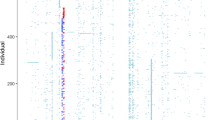
The GABRG1 genomic region, including 5 kb upstream and 1 kb downstream, was retrieved from NCBI Human Genome Build 35.1. Genotype data from the most diverse population, Africans, were obtained from the HapMap Project Public Release no. 18 to construct the GABRG1 haplotype structure using SNPHAP (http://www-gene.cimr.cam.ac.uk/clayton/software/snphap.txt). To select minimum index SNPs that represent maximal haplotype structures information, a program based on a double classification tree search algorithm was used (Zhang et al, 2004). On this basis, 24 SNPs spanning GABRG1 were selected (Figure 1). Genotyping was performed using SNPlex (Tobler et al, 2005). Allele-specific probes and optimized multiplexed assays for the 24 SNPs were designed by an automated multistep pipeline (Applied Biosystems, ABI: http://www.appliedbiosystems.com). SNPlex was carried out on fragmented gDNA (88 ng per well) according to the manufacturer's protocols. Samples were run on the 3730 DNA Analyzer (ABI) and data were analyzed using Gene Mapper version 4.0 software (ABI). Genotype analysis was performed based on the SNPlex_Rules_3730 method following the factory default rules. Genotyping included 21% duplicates.
GABRG1 gene map and haplotype block structure in HapMap populations. The positions of the genotyped SNPs are indicated. CDS=coding sequences, linked by dotted lines to the corresponding exons. The haplotype block structures of the three HapMap populations: CEU (Caucasian); YRI (African), and ASN (Chinese/Japanese) are provided. Conservation across 17 vertebrate species, from zebrafish to humans is indicated.
To determine the haplotype block structure across the 493 kb GABRG1 and GABRA2 region, 10 GABRA2 SNPs from the Illumina Linkage IV-B panel, previously genotyped with the Illumina GoldenGate platform under contract at Illumina (Hodgkinson et al, 2008), were included in the analyses. In addition, two previously genotyped GABRA2 SNPs (rs279863 and rs279858; Enoch et al, 2006a) that tagged the two common GABRA2 yin-yang haplotypes, were included. These two SNPs, particularly rs279858, have been most strongly associated with alcohol dependence in earlier studies. The inclusion of these two SNPs allows for cross comparison with earlier studies. The genotyped GABRA2 SNPs were (5′–3′): rs11503014; haplotype block 1: rs9291283, rs1442060; haplotype block 2: rs10805145, rs426463, rs279827, rs279843, rs279847, rs279858, rs279863, rs519270, and rs693547.
Assessment of Population Stratification using Ancestry Informative Markers (AIMs)
The Plains Indian and Finnish Caucasian samples were genotyped for 186 ancestry markers (Hodgkinson et al, 2008). These AIMs were also genotyped in 1051 individuals from the 51 worldwide populations represented in the HGDP-CEPH Human Genome Diversity Cell Line Panel (http://www.cephb.fr/HGDP-CEPH-Panel). PHASE Structure 2.2 (http://pritch.bsd.uchicago.edu/software.html) was run, simultaneously using the AIMs genotypes from our two samples and the 51 CEPH populations to identify population substructure and compute individual ethnic factor scores. This ancestry assessment showed that within the Plains Indian sample there was on average 0.05 admixture with Caucasians (median value=0.01) and 0.01 with African Americans. In the Finnish Caucasians, the only detectable admixture was with an Asian factor (mean=0.07, median=0.02). Because the ethnic factor scores were highly skewed (due to very low admixture) we used a median split in analyses. Ethnic factor scores were initially tested as covariates in the Plains Indian and Finnish Caucasian analyses but their inclusion had no significant effects on any analyses.
Statistical Analyses
In both the Plains Indian and the Finnish Caucasian samples, the percentage of genetic identity shared between any two individuals through common descent was calculated for all possible pairs (related and unrelated) using SAGE (Case Western Reserve University). The average sharing of descent was 0.003 in the Plains Indians, which is less than the degree of relationship between third cousins. The average sharing of descent between any two Finnish Caucasian individuals was only 0.002.
As the Plains Indians derive from one large pedigree we confirmed the significant p-values for associations by performing 10 000 simulations using SIMPED (Leal et al, 2005). With this program genotypes can be generated conditional on user-specified frequencies and using the gene-drop method within pedigrees of virtually any size and complexity. As most of the significant SNPs were in allelic identity, association analysis (t-test between the two homozygous genotypes) was computed for the simulated SNP rs12511372 and a null distribution for the t-statistic was generated. This was subsequently used to determine the empirical P-value (0.0004), which was close to the p-value assuming independence (0.0003). Therefore we were able to undertake analyses that assume independence of individuals.
Haplotype frequencies were estimated using a Bayesian approach implemented with PHASE (Stephens and Donnelly, 2003). Haploview version 2.04 software (Whitehead Institute for Biomedical Research, USA) was used to produce LD matrices with D′ set to 0.80.
Many of the GABRG1 SNPs were in allelic identity (Table 1). Twelve SNPs were selected to capture all the allelic variation across GABRG1 and the intergenic regions. The two GABRA2 SNPs that tag the GABRA2 risk haplotypes were included. PHASE was employed to determine long-distance haplotypes across the two gene regions using the 14 SNPs indicated in Table 1.
RESULTS
GABRG1 SNP Associations
Allele frequencies in both samples were very similar across GABRG1 however they began to differ in the intergenic region and differed markedly in GABRA2 (Table 1). In both the Plains Indians and the Finns, GABRG1 SNPs extending from intron 1 to downstream from the 3-UTR region were significantly associated with AUD (Table 1). In the Finns, SNPs in the GABRG1 promoter and the intergenic region between GABRG1 and GABRA2 were also associated with AUD. The allelic associations were in the same direction in both samples.
GABRG1 Haplotype Associations
Plains Indians. There was one haplotype block extending from rs12511372 in intron 1 of GABRG1 to rs1504501 distal to the gene, a distance of 155 kb (Figure 2). Twelve of the 16 SNPs within the haplotype block showed significant associations with AUD (Table 1). There were seven haplotypes with ⩾0.01 frequency that accounted for 0.93 of the haplotype diversity. From Figure 2 it can be seen that there are two cladistic groupings of haplotypes: H1–H4 and H5–H7. Haplotypes H1–H4 were more common in alcoholics than non-alcoholics (0.54 vs 0.41). The other three haplotypes (H5–H7) were more abundant in non-alcoholics compared with alcoholics (0.59 vs 0.46): H1–H4 vs H5–H7: χ2=8.8, 1 d.f., p=0.003; global analysis: χ2=21.4, 6 d.f., p=0.002.
GABRG1 haplotype block structure and haplotypes in Plains Indians. (a) The haplotype block extends from rs12511372 in intron 1 to rs1504501 distal to the gene (155 kb). The darker the color of squares, the greater the degree of linkage disequilibrium (D′). (b) Cladistic groupings of haplotypes with frequency ⩾0.01 in total sample: increased frequency of group H1–H4 in alcoholics; increased frequency of group H5–H7 in non-alcoholics; p=0.003. Alleles 1 and 2 are located on opposite DNA strands.
Finnish Caucasians. There were two GABRG1 haplotype blocks with a recombination region in intron 2 (Figure 3). Block 1 extended from the GABRA2–GABRG1 intergenic SNP rs4695146 to the intron 2 SNP rs17536211, a distance of 83.2 kb. There were nine haplotypes (⩾0.01 frequency) that accounted for 0.96 of the haplotype diversity. From Figure 3 it can be seen that haplotypes H1 and H2 differed by only one allele and were more abundant in alcoholics than non-alcoholics (0.45 vs 0.36). In contrast, haplotypes H8 and H9, differing by only one allele, were more common in non-alcoholics compared with alcoholics (0.49 vs 0.41): H1+H2 vs H8+H9: χ2=8.3, 1 d.f., p=0.0039.
GABRG1 haplotype block structure and haplotypes in Finnish Caucasians. Haplotype block 1 extends 83 kb from the intergenic SNP rs4695146 to the intron 2 SNP rs17536211. Haplotype block 2 extends 114 kb from intron 2 SNP 13130508 to the downstream SNP rs1394344. The darker the color of squares, the greater the degree of linkage disequilibrium (D′). Cladistic groupings of haplotypes with frequency ⩾0.01 in total sample: block 2, increased frequency of group H1–H3 in alcoholics; increased frequency of group H4–H6 in non-alcoholics; p=0.005. Block 1, increased frequency of group H1–H2 in alcoholics; increased frequency of group H8–H9 in non-alcoholics; p=0.004. Alleles 1 and 2 are located on opposite DNA strands.
Block 2 extended from the intron 2 SNP rs13130508 to the intergenic SNP rs1394344, a distance of 113.7 kb. There were six haplotypes with a frequency of ⩾ 0.01 that accounted for 0.98 of the haplotype diversity. From Figure 3 it can be seen that haplotypes could be cladistically arranged into two groups: H1–H3, that were more abundant in alcoholics (0.52 vs 0.43) and H4–H6 that were more common in non-alcoholics (0.57 vs 0.48): H1–H3 vs H4–H6: χ2=8.1, 1 d.f., p=0.0045.
GABRA2 SNP and Haplotype Associations
Three SNPs, rs494270 (intergenic) and the GABRA2 tag SNPs rs279863 and rs279858, were located within the GABRA2 haplotype block 2 that has been associated with AUD in previous studies (Table 1). In the Finns SNPs rs279863 and rs279858, which are in allelic identity, showed a significant genotypic association with AUD due to the fact that alcoholics had an excess of both homozygotes: 11 (0.44 vs 0.34) and 22 (0.18 vs 0.16), and a corresponding deficit of heterozygotes (0.38 vs 0.50) compared with non-alcoholics, respectively. There were no significant differences in allele frequencies.
The Plains Indians had three common haplotypes that accounted for 0.99 of the haplotype diversity: 122 (0.64), 211 (0.25), and 111 (0.10). The Finnish Caucasians had only two haplotypes that accounted for 0.93 of the haplotype diversity: 211 (0.57) and 122 (0.36). There was no difference in haplotype frequency distribution between alcoholics and non-alcoholics in the Plains Indians (p=0.68, 2 d.f.) and the Finns (p=0.39, 1 d.f.).
Long-Distance Haplotypes Across GABRG1–GABRA2: Influence of GABRG1 on Association with AUD
Figure 4 shows that in the Finnish Caucasians and Plains American Indians there is a degree of LD extending from GABRA2 block 2 to downstream of GABRG1. For example, D′ between rs1504501 (in the intergenic region distal to GABRG1) and GABRA2 block 2 SNPs was 0.6 in the Finns and 0.4 in the Plains Indians.
A total of 14 SNPs (12 GABRG1 and 2 GABRA2, identified in Table 1) were included in haplotype analyses across GABRG1 and GABRA2 block 2, a distance of more than 354 kb, to determine extended haplotypes.
Finnish Caucasians. There were 13 long-distance haplotypes with ⩾0.01 frequency that accounted for 0.89 of the haplotype diversity. Of these, five haplotypes had a frequency of ⩾0.05 (H1, 0.30; H2, 0.07; H3, 0.11; H4, 0.16; and H5, 0.07), accounting for 0.71 of the haplotype diversity in the total sample. For simplicity, analyses were conducted with the haplotypes ⩾0.05 frequency. Analyses undertaken with these five haplotypes showed that H1 was more abundant in alcoholics, H3–H5 were more common in non-alcoholics and H2 was neutral: H1 vs rest: χ2=7.3, 1 d.f., p=0.007 (Figure 5).
The association of long-distance haplotypes across GABRG1 and GABRA2 with alcohol use disorders. See Table 1 for the 14 SNPs used to derive these long-distance haplotypes that are at least 341 kb long. There are three haplotypes (H1, H3, and H4) that are identical in both populations. H1 is associated with alcohol use disorders (AUD) in both populations. H3 and H4 are more abundant in non-alcoholics from both populations. Upward arrow indicates increased haplotype frequency in alcoholics, downward arrow indicates decreased frequency in alcoholics, ‘=’ indicates no association with AUD. Arrows and ‘=’ on left indicate direction of GABRG1 haplotypes' association with AUD. Arrows and ‘=’ on right indicate the direction of GABRG1–GABRA2 long-distance haplotypes' association with AUD. Alleles 1 and 2 are located on opposite DNA strands.
Figure 5 illustrates the relative contributions from GABRG1 and GABRA2 haplotypes to the extended haplotypes' association with AUD. H1: both the GABRG1 block 1 and block 2 haplotypes are increased in alcoholics, therefore extended haplotype H1 is increased in alcoholics. H2: the GABRG1 block 2 haplotype is increased in alcoholics, the block 1 haplotype is decreased in alcoholics therefore extended haplotype H2 is neutral to AUD diagnosis. Haplotypes H3 and H4 are less abundant in alcoholics: in both cases the GABRG1 block 1 and block 2 haplotypes are decreased in alcoholics. Haplotype H5 is numerically increased in non-alcoholics: the GABRG1 block 2 haplotype is neutral; the block 1 haplotype is more common in non-alcoholics. As discussed above, GABRA2 haplotypes were not associated with AUD and, as seen in Figure 5, did not contribute to the extended haplotypes' association with AUD.
Plains Indians. Nine haplotypes had a frequency of ⩾0.01 and accounted for 0.90 of the haplotype diversity. Of these, six haplotypes had a frequency of ⩾0.05: H1, 0.16; H2, 0.09; H5, 0.18; H6, 0.22; H3, 0.06; H4, 0.09, accounting for 0.80 of haplotype diversity in the total sample. For simplicity, analyses were conducted with the haplotypes ⩾0.05 frequency. As can be seen from Figure 5, there are two cladistic groupings of haplotypes, (H1, H2, and H5) and (H3, H4, and H6). Haplotypes H1, H2, and H5 were more abundant in alcoholics compared with non-alcoholics (0.58 vs 0.43); haplotypes H3, H4, and H6 were more common in non-alcoholics (0.57 vs 0.42): χ2=10.4, 1 d.f., p=0.0013.
Only one GABRG1 haplotype contributes to H1, H2, and H5: this GABRG1 haplotype is associated with AUD. The one GABRG1 haplotype that contributes to H6, H3, and H4 is more abundant in non-alcoholics. In contrast, the GABRA2 yin-yang haplotypes 211 and 122 are found in the long-distance haplotypes that are associated both with and without AUD; likewise for the intergenic haplotypes 111 and 222.
Ancestral Haplotypes
The Finns and Plains Indians have in common three long-distance haplotypes: H1, H3, and H4 (see Figure 5). These ancestral haplotypes accounted for 0.57 of the long-distance haplotype diversity in Finns and 0.31 of diversity in the Plains. In both populations, H1 is associated with AUD and H3 and H4 are more abundant in non-alcoholics (Figure 5). Haplotypes H3 and H4 have very nearly opposite configuration to haplotype H1.
Long-Distance Haplotypes Across GABRG1–GABRA2: Influence of GABRA2 on Association with AUD
Two GABRG1 SNPs that showed exceptionally high LD across the whole GABRG1–GABRA2 region in both samples: rs13130508 (D′=0.9–1.0) and rs10517150 (D′=0.9–1.0) were included in the group of 14 SNPs (Table 1) selected to detect extended haplotypes.
In the Finns, the variant alleles of these two GABRG1 SNPs were found on three extended haplotypes (total frequency=0.06) that only differed in the GABRA2 block 2 SNPs: haplotypes 22111221121122 (0.01); 22111221121211 (0.02); and 22111221121222 (0.03). These haplotypes differed in their association with AUD. Two haplotypes were more common in alcoholics than in non-alcoholics: 22111221121122 (0.29 vs 0.12) and 22111221121222 (0.57 vs 0.44). In contrast, 22111221121211 was more common in non-alcoholics compared with alcoholics (0.44 vs 0.14); χ2=6.8, 2 d.f., p=0.034.
In the Plains Indians, the variant alleles of the GABRG1 SNPs rs13130508 and rs10517150 were found on only one extended haplotype: 22111221222122 (frequency 0.04) that was not the same as any of the Finnish haplotypes.
DISCUSSION
We found substantial, significant GABRG1 haplotype and SNP associations with AUD in two independent populations. The 10 SNPs that showed significant associations with AUD were in approximate allelic identity in both populations (non-alcoholics: Plains Indians, MAF=0.40–0.46; Finns, MAF=0.35–0.50) suggesting selection pressure. In the Plains Indians the significant SNP associations were located within the one haplotype block in which GABRG1 (excluding the promoter region) is located. However, in the Finns nearly all the SNPs were associated with AUD in a region that extended from the distal intergenic region, across GABRG1 and up to the GABRA2 haplotype block. This suggests that the functional locus may be found within a long-distance haplotype. Moreover, the examination of more population isolates might help to narrow down the genomic interval in which the functional variant(s) resides.
In contrast to our findings with GABRG1 we did not find a GABRA2 haplotype association with AUD in either sample. In a previous analysis in these two populations we had found a complex GABRA2 association with AUD in men only that was mediated by trait anxiety (Enoch et al, 2006a). The observed findings in our current study suggest that the association signal coming from GABRG1 is more robust than that from GABRA2.
The Finnish Caucasian and Plains Indian haplotype block structure and extended LD patterns across GABRG1 and GABRA2 were similar to that found in the HapMap European American and Asian populations and in a recent study of 13 SNPs covering the region between GABRG1 intron 7 and GABRA2 intron 3 (Ittiwut et al, 2008). It has been shown that LD between distant markers is due to the presence of extended haplotype superblocks in individuals with ancient chromosomes which have escaped historic recombination (Buzas et al, 2004). When we looked at extended haplotypes with frequencies ⩾0.05, we found three long-distance haplotypes that were common to both populations with total frequencies that were lower in the Plains Indians (0.31) than in the Finns (0.57). The Plains Indians experienced a bottleneck in the recent genetic past therefore lower frequencies of ancestral haplotypes might be expected. The ancient superblock structure has been partially disrupted by recombination in the region between GABRG1 and GABRA2, and also in the Caucasian GABRG1 intron 2.
The most common of the three ancestral, extended haplotypes was associated with AUD in both populations; the other two ancestral haplotypes (identical except for the GABRA2 contribution) were more abundant in non-alcoholics. Our results suggest that the signal for the extended haplotypes' association with AUD derives from GABRG1 and is independent of GABRA2 (Figure 5). However, the differential impact of GABRA2 haplotypes was observed in the less common (<0.05 frequency) long-distance haplotypes. We had noted that in both samples, two GABRG1 SNPs, rs13130508 and rs10517150 (not associated with AUD), showed exceptionally high LD (D′=0.9–1.0) with GABRA2 SNPs indicating the likelihood that they are located in ancient chromosomal regions. However, the variant alleles of these two SNPs were not embedded in the three common ancestral haplotypes described above. Instead, in the Finns they were found in three haplotypes that differed only in the GABRA2 haplotype contribution and it was these GABRA2 haplotypes that determined the haplotype association with AUD. These three ancestral haplotypes account for only 0.06 of the extended haplotype diversity and therefore they are unlikely to contribute significantly to the previously reported AUD associations with the two common yin-yang GABRA2 haplotypes in Caucasians (Agrawal et al, 2006; Covault et al, 2004; Edenberg et al, 2004; Enoch et al, 2006a; Fehr et al, 2006; Lappalainen et al, 2005; Soyka et al, 2008).
Edenberg et al (2004) genotyped six GABRG1 SNPs in the family-based COGA data set extending from the 5′ region to intron 8. Their strongest association with alcohol dependence was with an intron 1 SNP rs1391175 (p=0.05). We genotyped two of these six SNPs: for the intron 1 SNP rs2221020 Edenberg et al found a trend association with alcohol dependence (p=0.07) in contrast to our significant associations with AUD in both populations (p<0.01). For the intron 8 SNP rs1497570, Edenberg et al found no association (p=0.65) in contrast to our significant results in both samples (p<0.05). Conversely, as in our study Covault et al (2008) have shown significant haplotype and SNP association with alcoholism in a haplotype block that extends from GABRG1 intron 2 to the intergenic region between GABRG1 and GABRA2 in two large groups of US Caucasians. They genotyped 15 SNPs extending from GABRA2 intron 3 to GABRG1 intron 3 and found, as we did, that although the GABRA2 SNPs were located in a haplotype block distinct from GABRG1 SNPs, there was moderate LD between markers in the two blocks. Covault et al (2008) used only 4 SNPs to identify GABRG1 haplotypes in contrast to our 11 SNPs, nevertheless both our studies have identified GABRG1 haplotype associations with alcoholism. Our study expanded on this earlier result by demonstrating in two independent, ethnically diverse populations that the association between AUD and GABRG1 extended across a region that included all of GABRG1 and the downstream intergenic region.
As in our study, Covault et al (2008) found that in the extended haplotypes, the AUD risk was conferred by the GABRG1 and not the GABRA2 haplotype components. Analyses of a best-fit genetic model for markers in the two haplotype block regions led Covault et al (2008) to conclude that there may be a separate contribution to risk for AD by GABRG1 and GABRA2 and that their earlier findings of a GABRA2 association with alcoholism (Covault et al, 2004) might be partly due to LD with functional genetic variants in GABRG1.
As yet, there is no known functional locus in either GABRA2 or GABRG1. Although GABRA2 has four common isoforms (Tian et al, 2005) GABRG1 has no known isoforms. There is a microRNA binding site in the GABRG1 3′ UTR, however no SNP overlaps with that site. The fact that the chr 4 genes cluster together and that there is extensive LD across the cluster suggests the presence of distant enhancers and repressors. Using NCBI and HapMap we determined that the nearest gene, GNPDA2 (glucosamine-6-phosphate deaminase 2), is at a distance of more than 1 Mb and several major haplotype blocks downstream from GABRG1. Moreover, long-distance LD does not extend downstream from the GABRG1 haplotype block. Therefore, it seems likely that the functional locus is located in the, admittedly extensive, region covered by the extended haplotypes. We used HapMap haplotypes for the Caucasian, Asian, and African samples to determine the two major ancestral GABRG1 haplotypes (H1 and H3+H4, as shown in Figure 5) and created the haplotype-specific sequences from NCBI refGene's GABRG1 transcript (NM_173536.3). We ran Mfold 3.2 (Zuker et al, 1999) with the haplotype-specific transcript sequences and computed 10 different foldings for each haplotype. There were no significant differences in the folding energy between the two ancestral haplotypes (p=0.187).
GABRG1 has until recently been in the shadow of its neighbor, GABRA2, and has not been considered a prime candidate gene for alcoholism. In the rat, GABRG1 is expressed in only a few brain regions, primarily in the amygdala and all areas receiving innervation from the striatum including the substantia nigra (Pirker et al, 2000; Schwarzer et al, 2001). These are regions that are implicated in reward and addiction. The GABAA receptor subunit composition determines distinct pharmacological and electrophysiological properties. Studies have shown that γ1 subunits co-assemble with α2 subunits in vivo largely as α2β1γ1 receptors (Whiting, 2003) that are much less responsive to the effects of benzodiazepines than receptors with the much more common γ2 subunit (Wafford et al, 1993). In contrast, the anxiolytic effects of benzodiazepines and barbiturates appear to be mediated in part by α2 subunits co-assembled with γ2 subunits (Dixon et al, 2008; Low et al, 2000). Thus it is possible that α2β1γ1 receptors may be implicated in alcoholism vulnerability per se whereas variation in the more common receptors with co-assembled α2 and γ2 subunits may predispose to alcoholism mediated by anxiety.
In conclusion, our similar results in two independent populations, Plains Indians and Finnish Caucasians, have identified a complex situation: (1) GABRG1 haplotypes and SNPs are significantly associated with AUD; (2) GABRA2 haplotypes are not associated with AUD except, as previously shown (Enoch et al, 2006a), when mediated by trait anxiety; (3) there are three common ancestral haplotypes that span GABRG1 and GABRA2; their association with AUD is determined by GABRG1 (4) in the Finns, the association of three less common (<0.05) long-distance haplotypes with AUD is determined by GABRA2. Our results suggest that there are likely to be independent contributions from both GABRG1 and GABRA2 to the risk for AUD.
References
Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL et al (2006). Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav Genet 36: 640–650.
American Psychiatric Association (1987). Diagnostic and Statistical Manual of Mental Disorders, 3rd (edn), revised American Psychiatric Association Press: Washington, DC.
Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G et al (1998). International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev 50: 291–313.
Buzas B, Belfer I, Hipp H, Lorincz I, Evans C, Phillips G et al (2004). Haplotype block and superblock structures of the alpha1-adrenergic receptor genes reveal echoes from the chromosomal past. Mol Genet Genomics 272: 519–529.
Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR (2004). Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet 129B: 104–109.
Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR (2008). Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology 33: 837–848.
Dixon CI, Rosahl TW, Stephens DN (2008). Targeted deletion of the GABRA2 gene encoding alpha2-subunits of GABA(A) receptors facilitates performance of a conditioned emotional response, and abolishes anxiolytic effects of benzodiazepines and barbiturates. Pharmacol Biochem Behav 90: 1–8.
Drgon T, D'Addario C, Uhl GR (2006). Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions. Am J Med Genet B Neuropsychiatr Genet 141: 854–860.
Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO et al (2004). Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet 74: 705–714.
Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D (2006a). Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am J Med Genet B Neuropsychiatr Genet 141: 599–607.
Enoch M-A, Waheed J, Harris CR, Albaugh B, Goldman D (2006b). Sex differences in the influence of COMT val158met on alcoholism and smoking in plains American Indians. Alcohol Clin Exp Res 30: 399–406.
Enoch M-A (2008). The role of GABA(A) receptors in the development of alcoholism. Pharmacol Biochem Behav 90: 95–104.
Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C et al (2006). Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet 16: 9–17.
Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA et al (2008). Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol 43: 505–515.
Ittiwut C, Listman J, Mutirangura A, Malison R, Covault J, Kranzler HR et al (2008). Interpopulation linkage disequilibrium patterns of GABRA2 and GABRG1 genes at the GABA cluster locus on human chromosome 4. Genomics 91: 61–69.
Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA et al (2006). Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Arch Gen Psychiatry 63: 957–968.
Lappalainen J, Long JC, Eggert M, Ozaki N, Robin RW, Brown GL et al (1998). Linkage of antisocial alcoholism to the serotonin 5-HT1B receptor gene in 2 populations. Arch Gen Psychiatry 55: 989–994.
Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E et al (2005). Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res 29: 493–498.
Leal SM, Yan K, Muller-Myhsok B (2005). SimPed: a simulation program to generate haplotype and genotype data for pedigree structures. Hum Hered 60: 119–122.
Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E et al (1998). Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet 81: 216–221.
Low K, Crestani F, Keist R, Benke D, Brunig I, Benson JA et al (2000). Molecular and neuronal substrate for the selective attenuation of anxiety. Science 290: 131–134.
Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY (2007). The Role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. J Stud Alcohol Drugs 68: 625–633.
Okada H, Matsushita N, Kobayashi K, Kobayashi K (2004). Identification of GABAA receptor subunit variants in midbrain dopaminergic neurons. J Neurochem 89: 7–14.
Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, Sperk G (2000). GABA(A) receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience 101: 815–850.
Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P et al (1998). Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81: 207–215.
Schwarzer C, Berresheim U, Pirker S, Wieselthaler A, Fuchs K, Sieghart W et al (2001). Distribution of the major gamma-aminobutyric acid(A) receptor subunits in the basal ganglia and associated limbic brain areas of the adult rat. J Comp Neurol 433: 526–549.
Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B (2008). GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res 42: 184–191.
Steiger JL, Russek SJ (2004). GABAA receptors: building the bridge between subunit mRNAs, their promoters, and cognate transcription factors. Pharmacol Ther 101: 259–281.
Stephens M, Donnelly P (2003). A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73: 1162–1169.
Tian H, Chen HJ, Cross TH, Edenberg HJ (2005). Alternative splicing and promoter use in the human GABRA2 gene. Brain Res Mol Brain Res 137: 174–183.
Tobler A, Short S, Andersen M, Paner T, Briggs J, Lambert S et al (2005). The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech 16: 398–406.
Wafford KA, Bain CJ, Whiting PJ, Kemp JA (1993). Functional comparison of the role of gamma subunits in recombinant human gamma-aminobutyric acidA/benzodiazepine receptors. Mol Pharmacol 44: 437–442.
Whiting PJ (2003). GABA-A receptor subtypes in the brain: a paradigm for CNS drug discovery? Drug Discov Today 8: 445–450.
Wisden W, Laurie DJ, Monyer H, Seeburg PH (1992). The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. I. Telencephalon, diencephalon, mesencephalon. J Neurosci 12: 1040–1062.
Zhang P, Sheng H, Uehara R (2004). A double classification tree search algorithm for index SNP selection. BMC Bioinformatics 5: 89.
Zinn-Justin A, Abel L (1999). Genome search for alcohol dependence using the weighted pairwise correlation linkage method: interesting findings on chromosome 4. Genet Epidemiol 17 (Suppl 1): S421–S426.
Zuker M, Mathews DH, Turner DH (1999). Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC (eds). RNA Biochemistry and Biotechnology, NATO ASI Series. Kluwer: Dordrecht, NL. pp 11–43.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institute on Alcohol Abuse and Alcoholism, NIH, and in part by the Office of Research on Minority Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
DISCLOSURE/CONFLICTS OF INTEREST
All authors reported no biomedical financial interests or potential conflicts of interest.
Rights and permissions
About this article
Cite this article
Enoch, MA., Hodgkinson, C., Yuan, Q. et al. GABRG1 and GABRA2 as Independent Predictors for Alcoholism in Two Populations. Neuropsychopharmacol 34, 1245–1254 (2009). https://doi.org/10.1038/npp.2008.171
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2008.171
Keywords
This article is cited by
-
Time-varying Effects of GABRG1 and Maladaptive Peer Behavior on Externalizing Behavior from Childhood to Adulthood: Testing Gene × Environment × Development Effects
Journal of Youth and Adolescence (2020)
-
GABAA receptor polymorphisms in alcohol use disorder in the GWAS era
Psychopharmacology (2018)
-
Endophenotypes for Alcohol Use Disorder: An Update on the Field
Current Addiction Reports (2015)
-
Association of Gamma-Aminobutyric Acid A Receptor α2 Gene (GABRA2) with Alcohol Use Disorder
Neuropsychopharmacology (2014)
-
Examination of Genetic Variation in GABRA2 with Conduct Disorder and Alcohol Abuse and Dependence in a Longitudinal Study
Behavior Genetics (2014)