Abstract
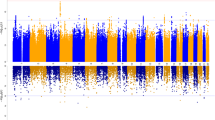
Obsessive-compulsive disorder (OCD) is a common, debilitating neuropsychiatric illness with complex genetic etiology. The International OCD Foundation Genetics Collaborative (IOCDF-GC) is a multi-national collaboration established to discover the genetic variation predisposing to OCD. A set of individuals affected with DSM-IV OCD, a subset of their parents, and unselected controls, were genotyped with several different Illumina SNP microarrays. After extensive data cleaning, 1465 cases, 5557 ancestry-matched controls and 400 complete trios remained, with a common set of 469 410 autosomal and 9657 X-chromosome single nucleotide polymorphisms (SNPs). Ancestry-stratified case–control association analyses were conducted for three genetically-defined subpopulations and combined in two meta-analyses, with and without the trio-based analysis. In the case–control analysis, the lowest two P-values were located within DLGAP1 (P=2.49 × 10−6 and P=3.44 × 10−6), a member of the neuronal postsynaptic density complex. In the trio analysis, rs6131295, near BTBD3, exceeded the genome-wide significance threshold with a P-value=3.84 × 10−8. However, when trios were meta-analyzed with the case–control samples, the P-value for this variant was 3.62 × 10−5, losing genome-wide significance. Although no SNPs were identified to be associated with OCD at a genome-wide significant level in the combined trio–case–control sample, a significant enrichment of methylation QTLs (P<0.001) and frontal lobe expression quantitative trait loci (eQTLs) (P=0.001) was observed within the top-ranked SNPs (P<0.01) from the trio–case–control analysis, suggesting these top signals may have a broad role in gene expression in the brain, and possibly in the etiology of OCD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kaplan HI, Sadock BJ . Study guide and self-examination review for Kaplan and Sadock’s synopsis of psychiatry. 6th edn. Williams & Wilkins: Baltimore, 1998, pp 541.
Karno M, Golding JM, Sorenson SB, Burnam MA . The epidemiology of obsessive-compulsive disorder in five US communities. Arch Gen Psychiatry 1988; 45: 1094–1099.
Ruscio AM, Stein DJ, Chiu WT, Kessler RC . The epidemiology of obsessive-compulsive disorder in the National Comorbidity Survey Replication. Mol Psychiatry 2010; 15: 53–63.
Blazer DG, Kessler RC, McGonagle KA, Swartz MS . The prevalence and distribution of major depression in a national community sample: the National Comorbidity Survey. Am J Psychiatry 1994; 151: 979–986.
Ayuso-Mateos J . Global Burden of obsessive-compulsive disorder in the year 2000. World Health Organization: Geneva, 2006.
Luxenburger H . Hereditat und Familientypus der Zwangsneurotiker. Arch Psychiatry 1930; 91: 590–594.
Lewis A . Problems of obsessional illness. Proc R Soc Med 1935; 29: 325–336.
Brown FW . Heredity in the Psychoneuroses (Summary). Proc R Soc Med 1942; 35: 785–790.
Rudin E . On the problem of compulsive disease with special reference to its hereditary relations]. Arch Psychiatr Nervenkr Z Gesamte Neurol Psychiatr 1953; 191: 14–54.
Kringlen E . Obsessional neurotics: a long-term follow-up. Br J Psychiatry 1965; 111: 709–722.
Rosenberg CM . Familial aspects of obsessional neurosis. Br J Psychiatry 1967; 113: 405–413.
Carey G, Goldsmith H, Tellegen A, Gottesman I . Genetics and personality inventories: The limits of replication with twin data. Behav Genet 1978; 8: 299–313.
Insel TR, Hoover C, Murphy DL . Parents of patients with obsessive-compulsive disorder. Psychol Med 1983; 13: 807–811.
Rasmussen SA, Tsuang MT . Clinical characteristics and family history in DSM-III obsessive-compulsive disorder. Am J Psychiatry 1986; 143: 317–322.
van Grootheest DS, Cath DC, Beekman AT, Boomsma DI . Twin studies on obsessive-compulsive disorder: a review. Twin Res Hum Genet 2005; 8: 450–458.
Iervolino AC, Rijsdijk FV, Cherkas L, Fullana MA, Mataix-Cols D . A multivariate twin study of obsessive-compulsive symptom dimensions. Arch Gen Psychiatry 2011; 68: 637–644.
Moore J, Smith GW, Shevlin M, O′Neill FA . Alternative factor models and heritability of the Short Leyton Obsessional Inventory-Children′s Version. J Abnorm Child Psychol 2010; 38: 921–934.
Bolton D, Rijsdijk F, Eley TC, O′Connor TG, Briskman J, Perrin S . Normative childhood repetitive routines and obsessive compulsive symptomatology in 6-year-old twins. J Child Psychol Psychiatry 2009; 50: 1139–1146.
Hanna GL, Veenstra-VanderWeele J, Cox NJ, Boehnke M, Himle JA, Curtis GC et al. Genome-wide linkage analysis of families with obsessive-compulsive disorder ascertained through pediatric probands. Am J Med Genet 2002; 114: 541–552.
Shugart YY, Samuels J, Willour VL, Grados MA, Greenberg BD, Knowles JA et al. Genomewide linkage scan for obsessive-compulsive disorder: evidence for susceptibility loci on chromosomes 3q, 7p, 1q, 15q, and 6q. Mol Psychiatry 2006; 11: 763–770.
Willour VL, Yao Shugart Y, Samuels J, Grados M, Cullen B, Bienvenu OJ et al. Replication study supports evidence for linkage to 9p24 in obsessive-compulsive disorder. Am J Hum Genet 2004; 75: 508–513.
Hanna GL, Veenstra-Vanderweele J, Cox NJ, Van Etten M, Fischer DJ, Himle JA et al. Evidence for a susceptibility locus on chromosome 10p15 in early-onset obsessive-compulsive disorder. Biol Psychiatry 2007; 62: 856–862.
Liang KY, Wang Y, Shugart YY, Grados M, Fyer AJ, Rauch S et al. Evidence for potential relationship between SLC1A1 and a putative genetic linkage region on chromosome 14q to obsessive-compulsive disorder with compulsive hoarding. Am J Med Genet B Neuropsychiatr Genet 2008; 147B: 1000–1002.
Ross J, Badner J, Garrido H, Sheppard B, Chavira DA, Grados M et al. Genomewide linkage analysis in Costa Rican families implicates chromosome 15q14 as a candidate region for OCD. Hum Genet 2011; 6: 795–805.
Samuels J, Shugart YY, Grados MA, Willour VL, Bienvenu OJ, Greenberg BD et al. Significant linkage to compulsive hoarding on chromosome 14 in families with obsessive-compulsive disorder: results from the OCD Collaborative Genetics Study. Am J Psychiatry 2007; 164: 493–499.
Pittenger C, Bloch MH, Williams K . Glutamate abnormalities in obsessive compulsive disorder: neurobiology, pathophysiology, and treatment. Pharmacol Ther 2011; 132: 314–332.
Wu K, Hanna GL, Rosenberg DR, Arnold PD . The role of glutamate signaling in the pathogenesis and treatment of obsessive-compulsive disorder. Pharmacol Biochem Behav 2011; 100: 726–735.
Stewart SE, Platko J, Fagerness J, Birns J, Jenike E, Smoller JW et al. A genetic family-based association study of OLIG2 in obsessive-compulsive disorder. Arch Gen Psychiatry 2007; 64: 209–214.
Stewart SE, Fagerness JA, Platko J, Smoller JW, Scharf JM, Illmann C et al. Association of the SLC1A1 glutamate transporter gene and obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet 2007; 144B: 1027–1033.
Arnold PD, Sicard T, Burroughs E, Richter MA, Kennedy JL . Glutamate transporter gene SLC1A1 associated with obsessive-compulsive disorder. Arch Gen Psychiatry 2006; 63: 769–776.
Dickel DE, Veenstra-VanderWeele J, Cox NJ, Wu X, Fischer DJ, Van Etten-Lee M et al. Association testing of the positional and functional candidate gene SLC1A1/EAAC1 in early-onset obsessive-compulsive disorder. Arch Gen Psychiatry 2006; 63: 778–785.
Wendland JR, Moya PR, Timpano KR, Anavitarte AP, Kruse MR, Wheaton MG et al. A haplotype containing quantitative trait loci for SLC1A1 gene expression and its association with obsessive-compulsive disorder. Arch Gen Psychiatry 2009; 66: 408–416.
Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 2007; 448: 894–900.
Züchner S, Wendland JR, Ashley-Koch AE, Collins AL, Tran-Viet KN, Quinn K et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol Psychiatry 2009; 14: 6–9.
Bienvenu OJ, Wang Y, Shugart YY, Welch JM, Grados MA, Fyer AJ et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am J Med Genet B Neuropsychiatr Genet 2009; 150B: 710–720.
Boardman L, van der Merwe L, Lochner C, Kinnear CJ, Seedat S, Stein DJ et al. Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Compr Psychiatry 2011; 52: 181–187.
Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry 2009; 166: 540–556.
Gamazon ER, Huang RS, Cox NJ, Dolan ME . Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci USA 2010; 107: 9287–9292.
Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–976.
Lee PH, O’Dushlaine C, Thomas B, Purcell SM . INRICH: interval-based enrichment analysis for genome wide association studies. Bioinformatics 2012; 28: 1797–1799.
American Psychiatric Association.. Diagnostic criteria from DSM-IV-TR. American Psychiatric Association: Washington, D.C., 2000. xii, pp 370.
Scharf J . Genome-wide association study of Tourette Syndrome. Mol Psychiatry 2012.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Willer CJ, Li Y, Abecasis GR . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Hemminger BM, Saelim B, Sullivan PF . TAMAL: an integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics 2006; 22: 626–627.
Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 2006; 34 (Database issue): D590–D598.
The International HapMap C. A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2007; 35 (Database issue): D5–12.
Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL et al. SCAN: SNP and copy number annotation. Bioinformatics 2010; 26: 259–262.
Saccone SF, Saccone NL, Swan GE, Madden PA, Goate AM, Rice JP et al. Systematic biological prioritization after a genome-wide association study: an application to nicotine dependence. Bioinformatics 2008; 24: 1805–1811.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM et al. The human genome browser at UCSC. Genome Res 2002; 12: 996–1006.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ . Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 2010; 6: e1000888.
Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010; 6: e1000952.
Howie BN, Donnelly P, Marchini J . A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5: e1000529.
Consortium GP . A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–1073.
Dudbridge F, Gusnanto A . Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008; 32: 227–234.
Torii M, Hashimoto-Torii K, Levitt P, Rakic P . Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature 2009; 461: 524–528.
Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G et al. Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol Psychiatry 2009; 14: 416–428.
Sabéran-Djoneidi D, Marey-Semper I, Picart R, Studler JM, Tougard C, Glowinski J et al. A 19-kDa protein belonging to a new family is expressed in the Golgi apparatus of neural cells. J Biol Chem 1995; 270: 1888–1893.
Schweitzer B, Suter U, Taylor V . Neural membrane protein 35/Lifeguard is localized at postsynaptic sites and in dendrites. Brain Res Mol Brain Res 2002; 107: 47–56.
Rivière JB, Xiong L, Levchenko A, St-Onge J, Gaspar C, Dion Y et al. Association of intronic variants of the BTBD9 gene with Tourette syndrome. Arch Neurol 2009; 66: 1267–1272.
Perez-Torrado R, Yamada D, Defossez PA . Born to bind: the BTB protein-protein interaction domain. Bioessays 2006; 28: 1194–1202.
Sud A, Del Bono EA, Haines JL, Wiggs JL . Fine mapping of the GLC1 K juvenile primary open-angle glaucoma locus and exclusion of candidate genes. Mol Vis 2008; 14: 1319–1326.
Stewart SE, Rosario MC, Brown TA, Carter AS, Leckman JF, Sukhodolsky D et al. Principal components analysis of obsessive-compulsive disorder symptoms in children and adolescents. Biol Psychiatry 2007; 61: 285–291.
NIMH Transcriptional Atlas of Human Brain Development. Accessed on 11 November 2011. www.developinghumanbrain.org.
Zhang P, Xiang N, Chen Y, Sliwerska E, McInnis MG, Burmeister M et al. Family-based association analysis to finemap bipolar linkage peak on chromosome 8q24 using 2500 genotyped SNPs and 15 000 imputed SNPs. Bipolar Disord 2010; 12: 786–792.
Wieczorek L, Maas Jr JW, Muglia LM, Vogt SK, Muglia LJ . Temporal and regional regulation of gene expression by calcium-stimulated adenylyl cyclase activity during fear memory. PLoS One 2010; 5: e13385.
Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR . Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol Med 2010; 16: 247–253.
Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012; 17: 142–153.
de Mooij-van Malsen AJ, van Lith HA, Oppelaar H, Hendriks J, de Wit M, Kostrzewa E et al. Interspecies trait genetics reveals association of Adcy8 with mouse avoidance behavior and a human mood disorder. Biol Psychiatry 2009; 66: 1123–1130.
Kantojärvi K, Onkamo P, Vanhala R, Alen R, Hedman M, Sajantila A et al. Analysis of 9p24 and 11p12-13 regions in autism spectrum disorders: rs1340513 in the JMJD2C gene is associated with ASDs in Finnish sample. Psychiatr Genet 2010; 20: 102–108.
Acknowledgements
The authors would like to express their utmost gratitude to the OCD-affected families who participated in this research. In addition, they would like to thank the International OCD Foundation (IOCDF) for their role in establishing the IOCDF Genetics Collaborative, as well as other individuals who played roles in assisting this study, including Rhonda Ellwyn, Katherine Beattie, Colm O’Dushlaine, Doug Ruderfer, Priya Moorjani and V. Guttenthaler. This work was supported primarily by a grant from the Judah Foundation (a private, non-industry related foundation established by a family affected by OCD), NIH grants MH079489 and MH073250 to DLP, American Recovery and Re-investment Act (ARRA) awards NS40024-07S1 and NS16648-29S1 to DLP, by an American Academy of Child and Adolescent Psychiatry (AACAP) Early Investigator Research Grant, an Anxiety Disorders Association of America (ADAA) Junior Investigator Research Grant, the University of British Columbia and a Michael Smith Foundation Clinical Research Scholar Award to SES, and grants from the Tourette Syndrome Association (DLP and JMS), the American Academy of Neurology Foundation (JMS) and NIH grant MH085057 to JMS. The Broad Institute Center for Genotyping and Analysis was supported by grant U54 RR020278 from the National Center for Research Resources. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392) and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for related genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). The data sets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p. Frontal lobe eQTL data was provided by the North American Brain Expression Consortium and the UK Human Brain Expression Database. Funding support for generation of the eQTL data was provided by the UK Medical Research Council and the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services project Z01 AG000932-02. The North American Brain Expression Consortium comprises: Sampath Arepalli, Mark R Cookson, Allissa Dillman, Luigi Ferrucci, J Raphael Gibbs, Dena G Hernandez, Robert Johnson, Dan L Longo, Michael A Nalls, Richard O′Brien, Andrew Singleton, Bryan Traynor, Juan Troncoso, Marcel van der Brug, H Ronald Zielke and Alan Zonderman. The UK Human Brain Expression Database membership comprises: John A Hardy, Mina Ryten, Colin Smith, Daniah Trabzuni, Robert Walker and Mike Weale. None of the funding sources supporting this work had any influence or played any role in: (a) the design or conduct of the study; (b) management, analysis or interpretation of the data; or c) preparation, review or approval of the manuscript.
AUTHOR CONTRIBUTIONS
Manuscript preparation: SE Stewart, JA Knowles, D Yu, JM Scharf, CA Mathews, PD Arnold, E Gamazon, PD Evans, GL Hanna, NJ Cox and DL Pauls. Study design: SE Stewart, JM Scharf, D Yu, JA Knowles, PD Arnold, CA Mathews, BM Neale, JA Fagerness, EH Cook, S Purcell, NJ Cox, G Nestadt and DL Pauls. Data analysis: D Yu, BM Neale, S Purcell, JM Scharf, PD Evans, ER Gamazon, A Tikhomirov, A Pluzhnikov, A Konkashbaev, LK Davis, D Posthuma, E Eskin, C Sabatti, CK Edlund, DV Conti, JA Knowles, NJ Cox. Project management: SE Stewart, JM Scharf, JA Fagerness, MA Jenike and DL Pauls. Sample management and processing: JA Fagerness, S Haddad, JM Scharf, J Crane, C Mayerfeld and DL Pauls. Genotyping: AT Crenshaw, MA Parkin and DB Mirel. Phenotype management: SE Stewart, L Osiecki, D Hezel, C Illmann, JM Scharf and DL Pauls. Case sample collection (ordered by numbers of submitted samples): University of Bonn, Germany: M Wagner, R Moessner (Site PI), P Falkai, W Maier, S Ruhrmann, H-J Grabe, L Lennertz. Italy: L Bellodi, MC Cavallini. Toronto, Canada/Wayne State collaborative: PD Arnold, MA Richter, EH Cook, Jr, JL Kennedy, D Rosenberg. University of Cape Town, South Africa: DJ Stein (Site PI), SMJ Hemmings, C Lochner. UCSF/ Costa Rica collaborative: CA Mathews (Site PI), A Azzam, DA Chavira, E Fournier, H Garrido, B Sheppard, P Umana. National Institute of Mental Health: DL Murphy, JR Wendland. Michigan: GL Hanna (Site PI), J Veenstra-VanderWeele. AMC, Netherlands: D Denys (Site PI), R Blom, D Deforce, F Van Nieuwerburgh, HGM Westenberg. Wurzburg Germany: S Walitza (Site PI), K Egberts, T Renner. Massachusetts General Hospital, Boston: DL Pauls (Site PI), C Illmann, SE Stewart, JM Scharf, SL Rauch. Brazil: EC Miguel (Site PI), C Cappi, AG Hounie, MC do Rosario, AS Sampaio, H Vallada. Mexico: H Nicolini (Site PI), N Lanzagorta, B Camarena. Paris, France: M Leboyer (Site PI), R Delorme. University of Southern California: MT Pato (Site PI), CN Pato, JA Knowles, E Voyiaziakis. VUMC, Netherlands: DC Cath (Site PI), P Heutink, D Posthuma, JH Smit. OCGS, Johns Hopkins collaborative: G Nestadt (Site PI), J Samuels, OJ Bienvenu, B Cullen, AJ Fyer, MA Grados, BD Greenberg, JT McCracken, MA Riddle, Y Wang. Yale University: JF Leckman (Site PI), M Bloch, C Pittenger, V Coric. United Arab Emirates: V Eapen. Iowa: DW Black. Control Sample Collection: University Medical Center, Utrecht: RA Ophoff, E Strengman. University of Bonn: R Moessner (Site PI), M Wagner, P Falkai, W Maier, S Ruhrmann, H-J Grabe, L Lennertz. Data Collection: Italian Control data: F Macciardi, D Cusi, M Turiel, F Frau. eQTL and mQTL data: C Liu. MR Cookson, JR Gibbs and A Singleton for the North American Brain Expression Consortium; J Hardy for the UK Human Brain Expression Database.
North American Brain Expression Consortium
S Arepalli1, MR Cookson1, A Dillman1, L Ferrucci2, JR Gibbs1,3, DG Hernandez1,3, R Johnson4, DL Longo5, MA Nalls1, R O’Brien6, A Singleton1, B Traynor1, J Troncoso6, M van der Brug1,7, HR Zielke4, A Zonderman8;
UK Human Brain Expression Database
JA Hardy3, M Ryten3, C Smith9, D Trabzuni3, R Walker9, Mike Weale10
1Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; 2Clinical Research Branch, National Institute on Aging, Baltimore, MD, USA; 3Department of Molecular Neuroscience, UCL Institute of Neurology, London, UK; 4NICHD Brain and Tissue Bank for Developmental Disorders, University of Maryland Medical School, Baltimore, MD, USA; 5Lymphocyte Cell Biology Unit, Laboratory of Immunology, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA; 6Brain Resource Center, Johns Hopkins University, Baltimore, MD, USA; 7ITGR Biomarker Discovery Group, Genentech, South San Francisco, CA, USA; 8Research Resources Branch, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; 9Department of Pathology, The University of Edinburgh, Edinburgh, UK and 10King’s College London, Department of Medical & Molecular Genetics, UK.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Competing interests
SE Stewart has received funding from the International OCD Foundation (IOCDF) and is a member of the IOCDF Scientific Advisory Committee. PD Arnold reports funding sources including the CIHR, NIH, Ontario Research Foundation, Ontario Brain Institute, DNA Genotek, and the McLaughlin Centre. AB Singleton served as an unpaid consultant to Teva Pharmaceuticals. HJ Grabe has received funds from the German Research Foundation; Federal Ministry of Education and Research Germany; speakers honoraria from Bristol-Myers Squibb, Eli Lilly, Novartis, Eisai, Boehringer Ingelheim; speaker and travel funds from Janssen-Cilag, Eli Lilly, Novartis, AstraZeneca, Lundbeck and SALUS-Institute for Trend-Research and Therapy Evaluation in Mental Health. R Moessner has been supported by the German Research Foundation (DFG) (grants Wa 731/6 and 731/4), and by the German Federal Ministry for Education and Research (BMBF grant 01GV0907). M Wagner has been supported by the German Research Foundation (DFG) (grants Wa 731/6 and 731/4), and by the German Federal Ministry for Education and Research (BMBF grant 01GV0907). MA Richter has received honoraria from Lundbeck and she is recipient of grant funding from Eli Lilly Canada, Ontario Mental Health Foundation and the Obsessive-Compulsive Foundation. DJ Stein has received research grants and/or consultancy honoraria from Abbott, Astrazeneca, Eli-Lilly, GlaxoSmithKline, Jazz Pharmaceuticals, Johnson & Johnson, Lundbeck, Orion, Pfizer, Pharmacia, Roche, Servier, Solvay, Sumitomo, Takeda and Tikvah. JR Wendland is now a full-time employee of F Hoffmann-La Roche J Veenstra-VanderWeele receives research funding from Seaside Therapeutics, Roche Pharmaceuticals and Novartis. GL Hanna, MT Pato and CN Pato receive NIH funding. K Egberts, T Renner and S Walitza received sample collection funding by DFG WA168/1-1. SL Rauch has received research funding from Cyberonics and Medtronic. JA Knowles is a recipient of grant funding from NIH and from NARSAD; he sits on the Scientific Advisory Committee for Next-Generation Sequencing of Life Technologies and is a technical advisor to SoftGenetics; he is on the Scientific Advisory Committee for Next-Generation Sequencing of Life Technologies and is a technical advisor to SoftGenetics. JF Leckman has been funded by the NIH, the TSA, Talecris Biotherapeutics, Klingenstein Third Generation Foundation, John Wiley and Sons, McGraw Hill and Oxford University Press. V Coric works for Bristol Myers-Squibb. DW Black has received NIH funding and support from AstraZeneca and Psysadon in addition to royalties from American Psychiatric Publishing and Oxford University Press. The remaining authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
Rights and permissions
About this article
Cite this article
Stewart, S., Yu, D., Scharf, J. et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry 18, 788–798 (2013). https://doi.org/10.1038/mp.2012.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2012.85
Keywords
This article is cited by
-
In search of environmental risk factors for obsessive-compulsive disorder: study protocol for the OCDTWIN project
BMC Psychiatry (2023)
-
Unraveling the mechanisms of deep-brain stimulation of the internal capsule in a mouse model
Nature Communications (2023)
-
Comparative neurogenetics of dog behavior complements efforts towards human neuropsychiatric genetics
Human Genetics (2023)
-
Epigenetic modifications and obsessive–compulsive disorder: what do we know?
Brain Structure and Function (2023)
-
Epigenome-wide DNA methylation in obsessive-compulsive disorder
Translational Psychiatry (2022)