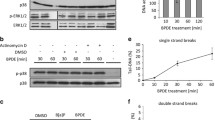
Abstract
Purpose
Environmental pollutants are known to induce DNA breaks, leading to genomic instability. Here, we propose a novel mechanism for the genotoxic effects exerted by environmentally exposed endocrine-disrupting chemicals (EDCs).
Methods
Bibliographic research and presentation of the analysis.
Discussion
In mammals, nucleotide excision repair, base excision repair, homologous recombination and non-homologous end-joining pathways are some of the major DNA repair pathways. p300 along with CREB-binding protein (CBP) contributes to chromatin remodeling, DNA damage response and repair of both single- and double-stranded DNA breaks. In addition to its role in DNA repair, CBP/p300 also acts as a coactivator to interact with the estrogen receptor and androgen receptor during its estrogen- and androgen-dependent transactivation, respectively. Since activated estrogen receptors (ERs) seize p300 from the repressed genes and redistribute it to the enhancer genes to activate transcription, the cellular functioning may be based on a balance between these pathways and any disturbance in one may alter the other, leading to undesirable physiological effects.
Conclusion
In conclusion, CBP/p300 is important for DNA repair and nuclear hormone receptor transactivation. Activated hormone receptors can sequester p300 to regulate the hormonal effects. Hence, we believe that activation of ERs by EDCs results in sequestration of CBP/p300 for ER transactivation and transcription initiation of its target genes, leading to a competition for CBP/P300, resulting in the deregulation of all other pathways involving p300/CBP.


Similar content being viewed by others
References
Damstra T, Barlow S, Bergman A, Kavlock R et al (2002) Global assessment of the state-of-the-science of endocrine disruptors. World Health Organization, Geneva, pp 11–32
De Coster S, van Larebeke N (2012) Endocrine-disrupting chemicals: associated disorders and mechanisms of action. J Environ Public Health 2012:713696
Foresta C, Tescari S, Di Nisio A (2018) Impact of perfluorochemicals on human health and reproduction: a male's perspective. J Endocrinol Invest 41:639–645
Bergman Å, Heindel JJ, Jobling S, Kidd K et al (2013) State of the science of endocrine disrupting chemicals 2012. World Health Organization, Geneva
Al-Saleh I, Al-Rajudi T, Al-Qudaihi G, Manogaran P (2017) Evaluating the potential genotoxicity of phthalates esters (PAEs) in perfumes using in vitro assays. Environ Sci Pollut Res Int 24:23903–23914
Ding ZM, Jiao XF, Wu D, Zhang JY et al (2017) Bisphenol AF negatively affects oocyte maturation of mouse in vitro through increasing oxidative stress and DNA damage. Chem Biol Interact 278:222–229
Ferreira LL, Couto R, Oliveira PJ (2015) Bisphenol A as epigenetic modulator: setting the stage for carcinogenesis? Eur J Clin Invest 45(Suppl 1):32–36
Prusinski Fernung LE, Yang Q, Sakamuro D, Kumari A et al (2018) Endocrine disruptor exposure during development increases incidence of uterine fibroids by altering DNA repair in myometrial stem cells. Biol Reprod 99:735–748
Li X, Yin P, Zhao L (2017) Effects of individual and combined toxicity of bisphenol A, dibutyl phthalate and cadmium on oxidative stress and genotoxicity in HepG 2 cells. Food Chem Toxicol 105:73–81
Maria VL, Correia AC, Santos MA (2003) Genotoxic and hepatic biotransformation responses induced by the overflow of pulp mill and secondary-treated effluents on Anguilla anguilla L. Ecotoxicol Environ Saf 55:126–137
Martinez-Paz P, Morales M, Martinez-Guitarte JL, Morcillo G (2013) Genotoxic effects of environmental endocrine disruptors on the aquatic insect Chironomus riparius evaluated using the comet assay. Mutat Res 758:41–47
Morales M, Martinez-Paz P, Ozaez I, Martinez-Guitarte JL et al (2013) DNA damage and transcriptional changes induced by tributyltin (TBT) after short in vivo exposures of Chironomus riparius (Diptera) larvae. Comp Biochem Physiol C Toxicol Pharmacol 158:57–63
Izzotti A, Kanitz S, D'Agostini F, Camoirano A et al (2009) Formation of adducts by bisphenol A, an endocrine disruptor, in DNA in vitro and in liver and mammary tissue of mice. Mutat Res 679:28–32
Santovito A, Cannarsa E, Schleicherova D, Cervella P (2018) Clastogenic effects of bisphenol A on human cultured lymphocytes. Hum Exp Toxicol 37:69–77
Iso T, Watanabe T, Iwamoto T, Shimamoto A et al (2006) DNA damage caused by bisphenol A and estradiol through estrogenic activity. Biol Pharm Bull 29:206–210
Fittipaldi S, Bimonte VM, Soricelli A, Aversa A et al (2019) Cadmium exposure alters steroid receptors and proinflammatory cytokine levels in endothelial cells in vitro: a potential mechanism of endocrine disruptor atherogenic effect. J Endocrinol Invest 42:727–739
Skipper A, Sims JN, Yedjou CG, Tchounwou PB (2016) Cadmium chloride induces DNA damage and apoptosis of human liver carcinoma cells via oxidative stress. Int J Environ Res Public Health 13:20
Guo X, Yang C, Qian X, Lei T et al (2013) Estrogen receptor alpha regulates ATM expression through miRNAs in breast cancer. Clin cancer Res 19:4994–5002
Pedram A, Razandi M, Evinger AJ, Lee E et al (2009) Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell 20:3374–3389
Song L, Lin C, Wu Z, Gong H et al (2011) miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE 6:e25454
Chen ST, Lin CC, Liu YS, Lin C et al (2013) Airborne particulate collected from central Taiwan induces DNA strand breaks, Poly(ADP-ribose) polymerase-1 activation, and estrogen-disrupting activity in human breast carcinoma cell lines. J Environ Sci Health A Tox Hazard Subst Environ Eng 48:173–181
Rebbeck TR (2000) Prophylactic oophorectomy in BRCA1 and BRCA2 mutation carriers. J Clin Oncol 18:100S–S103
He X, Jing Y, Wang J, Li K et al (2015) Significant accumulation of persistent organic pollutants and dysregulation in multiple DNA damage repair pathways in the electronic-waste-exposed populations. Environ Res 137:458–466
Jasso-Pineda Y, Diaz-Barriga F, Calderon J, Yanez L et al (2012) DNA damage and decreased DNA repair in peripheral blood mononuclear cells in individuals exposed to arsenic and lead in a mining site. Biol Trace Elem Res 146:141–149
De Flora S, Micale RT, La Maestra S, Izzotti A et al (2011) Upregulation of clusterin in prostate and DNA damage in spermatozoa from bisphenol A-treated rats and formation of DNA adducts in cultured human prostatic cells. Toxicol Sci 122:45–51
Dobrzynska MM, Radzikowska J (2013) Genotoxicity and reproductive toxicity of bisphenol A and X-ray/bisphenol A combination in male mice. Drug Chem Toxicol 36:19–26
Xin L, Wang J, Guo S, Wu Y et al (2014) Organic extracts of coke oven emissions can induce genetic damage in metabolically competent HepG2 cells. Environ Toxicol Pharmacol 37:946–953
Prasanth GK, Divya LM, Sadasivan C (2013) Bisphenol-A can inhibit the enzymatic activity of human superoxide dismutase. Hum Ecol Risk Assess 20:268–77
Nambiar M, Raghavan SC (2011) How does DNA break during chromosomal translocations? Nucleic Acids Res 39:5813–5825
Zhu C, Mills KD, Ferguson DO, Lee C et al (2002) Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109:811–821
Caldecott KW (2008) Single-strand break repair and genetic disease. Nat Rev Genet 9:619–631
Krokan HE, Bjoras M (2013) Base excision repair. Cold Spring Harb Perspect Biol 5:a012583
Scharer OD (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5:a012609
Dianov GL, Parsons JL (2007) Co-ordination of DNA single strand break repair. DNA Repair (Amst) 6:454–460
Chen J, Ghorai MK, Kenney G, Stubbe J (2008) Mechanistic studies on bleomycin-mediated DNA damage: multiple binding modes can result in double-stranded DNA cleavage. Nucleic Acids Res 36:3781–3790
Kryston TB, Georgiev AB, Pissis P, Georgakilas AG (2011) Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res 711:193–201
Nishana M, Raghavan SC (2012) Role of recombination activating genes in the generation of antigen receptor diversity and beyond. Immunology 137:271–281
Escargueil AE, Soares DG, Salvador M, Larsen AK et al (2008) What histone code for DNA repair? Mutat Res 658:259–270
Iizuka M, Smith MM (2003) Functional consequences of histone modifications. Curr Opin Genet Dev 13:154–160
Osley MA, Shen X (2006) Altering nucleosomes during DNA double-strand break repair in yeast. Trends Genet 22:671–677
Foulds CE, Feng Q, Ding C, Bailey S et al (2013) Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell 51:185–199
Vempati RK, Jayani RS, Notani D, Sengupta A et al (2010) p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem 285:28553–28564
Jang ER, Choi JD, Lee JS (2011) Acetyltransferase p300 regulates NBS1-mediated DNA damage response. FEBS Lett 585:47–52
Jain S, Wei J, Mitrani LR, Bishopric NH (2012) Auto-acetylation stabilizes p300 in cardiac myocytes during acute oxidative stress, promoting STAT3 accumulation and cell survival. Breast Cancer Res Treat 135:103–114
Ogiwara H, Ui A, Otsuka A, Satoh H et al (2011) Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30:2135–2146
Ogiwara H, Kohno T (2012) CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS ONE 7:e52810
Qi W, Chen H, Xiao T, Wang R et al (2016) Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis 31:193–203
Stauffer D, Chang B, Huang J, Dunn A et al (2007) p300/CREB-binding protein interacts with ATR and is required for the DNA replication checkpoint. J Biol Chem 282:9678–9687
Tillhon M, Cazzalini O, Nardo T, Necchi D et al (2012) p300/CBP acetyl transferases interact with and acetylate the nucleotide excision repair factor XPG. DNA Repair (Amst) 11:844–852
Cazzalini O, Sommatis S, Tillhon M, Dutto I et al (2014) CBP and p300 acetylate PCNA to link its degradation with nucleotide excision repair synthesis. Nucleic Acids Res 42:8433–8448
Hasan S, Hassa PO, Imhof R, Hottiger MO (2001) Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature 410:387–391
Wang QE, Han C, Zhao R, Wani G et al (2013) p38 MAPK- and Akt-mediated p300 phosphorylation regulates its degradation to facilitate nucleotide excision repair. Nucleic Acids Res 41:1722–1733
Hasan S, El-Andaloussi N, Hardeland U, Hassa PO et al (2002) Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol Cell 10:1213–1222
Hasan S, Stucki M, Hassa PO, Imhof R et al (2001) Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell 7:1221–1231
Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK et al (2006) Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol 26:1654–1665
Dutta A, Yang C, Sengupta S, Mitra S et al (2015) New paradigms in the repair of oxidative damage in human genome: mechanisms ensuring repair of mutagenic base lesions during replication and involvement of accessory proteins. Cell Mol Life Sci 72:1679–1698
Tini M, Benecke A, Um SJ, Torchia J et al (2002) Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 9:265–277
Likhite VS, Cass EI, Anderson SD, Yates JR et al (2004) Interaction of estrogen receptor alpha with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. J Biol Chem 279:16875–16882
Bannister AJ, Kouzarides T (1996) The CBP co-activator is a histone acetyltransferase. Nature 384:641–643
Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595–606
Kraus WL, Manning ET, Kadonaga JT (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol 19:8123–8135
Munshi N, Merika M, Yie J, Senger K et al (1998) Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell 2:457–467
Ogryzko VV, Schiltz RL, Russanova V, Howard BH et al (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953–959
Polesskaya A, Duquet A, Naguibneva I, Weise C et al (2000) CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem 275:34359–34364
Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S (2001) Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J Biol Chem 276:42753–42760
Boyes J, Byfield P, Nakatani Y, Ogryzko V (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594–598
Imhof A, Yang XJ, Ogryzko VV, Nakatani Y et al (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr Biol 7:689–692
Gronemeyer H, Gustafsson JA, Laudet V (2004) Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3:950–964
Rastinejad F, Huang P, Chandra V, Khorasanizadeh S (2013) Understanding nuclear receptor form and function using structural biology. J Mol Endocrinol 51:T1–T21
Safe S (2001) Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-p1 interactions. Vitam Horm 62:231–252
Marino M, Galluzzo P, Ascenzi P (2006) Estrogen signaling multiple pathways to impact gene transcription. Curr Genom 7:497–508
Sheppard HM, Harries JC, Hussain S, Bevan C et al (2001) Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol Cell Biol 21:39–50
Wong CW, Komm B, Cheskis BJ (2001) Structure-function evaluation of ER alpha and beta interplay with SRC family coactivators. ER selective ligands. Biochemistry 40:6756–6765
Demarest SJ, Martinez-Yamout M, Chung J, Chen H et al (2002) Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415:549–553
Stossi F, Madak-Erdogan Z, Katzenellenbogen BS (2009) Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol Cell Biol 29:1749–1759
Guertin MJ, Zhang X, Coonrod SA, Hager GL (2014) Transient estrogen receptor binding and p300 redistribution support a squelching mechanism for estradiol-repressed genes. Mol Endocrinol 28:1522–1533
Murakami S, Nagari A, Kraus WL (2017) Dynamic assembly and activation of estrogen receptor alpha enhancers through coregulator switching. Genes Dev 31:1535–1548
Acknowledgements
DLM is supported by Start-up Grant (YSS/2015/000987) from DST, Government of India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors hereby declare that they have no actual or potential competing/financial interest.
Ethical approval
This article doesn't involve any human subjects or animal work.
Informed consent
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lakshmanan, M.D., Shaheer, K. Endocrine disrupting chemicals may deregulate DNA repair through estrogen receptor mediated seizing of CBP/p300 acetylase. J Endocrinol Invest 43, 1189–1196 (2020). https://doi.org/10.1007/s40618-020-01241-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40618-020-01241-5