Abstract
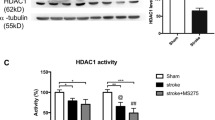
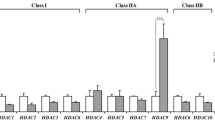
DNA methylation and histone acetylation can be modified by various pathological or physiological factors such as hypoxia, thus influencing gene expression. In this study, we investigated the changes of global DNA methylation and histone acetylation and the related enzymes in rat brain after chronic cerebrovascular hypoperfusion by bilateral common carotid occlusion (2-VO) surgery. Colorimetric and immunohistochemistry staining were used to evaluate the global DNA methylation and histone acetylation levels, respectively. The expressions of DNA methyltransferase 1/3a (DNMT1/3a), methyl-CpG binding domain protein 2 (MBD2), histone deacetylase 3 (HDAC3) and acetyltransferase (HAT) were assessed by Western blot. We found that the level of global DNA methylation was decreased to 31.7% (P <0.01) of the sham-operated group at 10 days and increased by 30% (P <0.01) compared with the sham group at 90 days after 2-VO surgery. DNMT3a expression was down-regulated to 75.7% of the sham group, while MBD2 expression was up-regulated by 95% compared with sham group at 90 days after 2-VO. The histone H3 acetylation level was markedly decreased to 75.3% of the sham group at 10 days and 73.5% at 90 days after 2-VO, while no significant change was found for histone H4 acetylation. HDAC3 expression was markedly down-regulated to 36% of the sham group, whereas cAMP-response element binding protein expression was up-regulated by 33.6% compared with the sham group at 90 days after 2-VO. These results suggest that chronic cerebrovascular hypoperfusion influences global DNA methylation and histone acetylation levels through the related enzymes, and therefore might contribute to several neurodegenerative diseases.
Similar content being viewed by others
References
Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol 2010, 90: 498–510.
Marques SC, Lemos R, Ferreiro E, Martins M, de Mendonca A, Santana I, et al. Epigenetic regulation of BACE1 in Alzheimer’s disease patients and in transgenic mice. Neuroscience 2012, 220: 256–266.
Scheen AJ, Junien C. Epigenetics, interface between environment and genes: role in complex diseases. Rev Med Liege 2012, 67: 250–257.
Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007, 447: 433–440.
Esteller M. Epigenetics in cancer. N Engl J Med 2008, 358: 1148–1159.
Gerhauser C. Cancer chemoprevention and nutriepigenetics: state of the art and future challenges. Top Curr Chem 2013, 329: 73–132.
Marques SC, Oliveira CR, Pereira CM, Outeiro TF. Epigenetics in neurodegeneration: a new layer of complexity. Prog Neuropsychopharmacol Biol Psychiatry 2011, 35: 348–355.
Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol 2009, 8: 1056–1072.
Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest 2005, 115: 1449–1457.
Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci 2005, 28: 195–204.
Hai J, Wan JF, Lin Q, Wang F, Zhang L, Li H, et al. Cognitive dysfunction induced by chronic cerebral hypoperfusion in a rat model associated with arteriovenous malformations. Brain Res 2009, 1301: 80–88.
Choi BR, Lee SR, Han JS, Woo SK, Kim KM, Choi DH, et al. Synergistic memory impairment through the interaction of chronic cerebral hypoperfusion and amlyloid toxicity in a rat model. Stroke 2011, 42: 2595–2604.
Almkvist O, Tallberg IM. Cognitive decline from estimated premorbid status predicts neurodegeneration in Alzheimer’s disease. Neuropsychology 2009, 23: 117–124.
Hai J, Lin Q, Li ST, Pan QG. Chronic cerebral hypoperfusion and reperfusion injury of restoration of normal perfusion pressure contributes to the neuropathological changes in rat brain. Brain Res Mol Brain Res 2004, 126: 137–145.
Farkas E, Luiten PG, Bari F. Permanent, bilateral common carotid artery occlusion in the rat: a model for chronic cerebral hypoperfusion-related neurodegenerative diseases. Brain Res Rev 2007, 54: 162–180.
Tatemichi TK, Desmond DW, Prohovnik I, Eidelberg D. Dementia associated with bilateral carotid occlusions: neuropsychological and haemodynamic course after extracranial to intracranial bypass surgery. J Neurol Neurosurg Psychiatry 1995, 58: 633–636.
Tsuda Y, Yamada K, Hayakawa T, Ayada Y, Kawasaki S, Matsuo H. Cortical blood flow and cognition after extracranialintracranial bypass in a patient with severe carotid occlusive lesions. A three-year follow-up study. Acta Neurochir (Wien) 1994, 129: 198–204.
Yang J, Ledaki I, Turley H, Gatter KC, Montero JC, Li JL, et al. Role of hypoxia-inducible factors in epigenetic regulation via histone demethylases. Ann N Y Acad Sci 2009, 1177: 185–197.
Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem 2008, 283: 36542–36552.
Xia X, Kung AL. Preferent ial binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol 2009, 10: R113.
Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat Res 2008, 640: 174–179.
Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics, the epicenter of the hypoxic response. Epigenetics 2010, 5: 293–296.
Ni J, Ohta H, Matsumoto K, Watanabe H. Progressive cognitive impairment following chronic cerebral hypoperfusion induced by permanent occlusion of bilateral carotid arteries in rats. Brain Res 1994, 653: 231–236.
Guo X, Wu X, Ren L, Liu G, Li L. Epigenetic mechanisms of amyloid-beta production in anisomycin-treated SH-SY5Y cells. Neuroscience 2011, 194: 272–281.
Liu H, Xing A, Wang X, Liu G, Li L. Regulation of betaamyloid level in the brain of rats with cerebrovascular hypoperfusion. Neurobiol Aging 2012, 33: 826 e831–842.
Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 1996, 87: 953–959.
Zhu H, Zhang J, Sun H, Zhang L, Liu H, Zeng X, et al. An enriched environment reverses the synaptic plasticity deficit induced by chronic cerebral hypoperfusion. Neurosci Lett 2011, 502: 71–75.
Shibata M, Yamasaki N, Miyakawa T, Kalaria RN, Fujita Y, Ohtani R, et al. Selective impairment of working memory in a mouse model of chronic cerebral hypoperfusion. Stroke 2007, 38: 2826–2832.
Wang X, Xing A, Xu C, Cai Q, Liu H, Li L. Cerebrovascular hypoperfusion induces spatial memory impairment, synaptic changes, and amyloid-beta oligomerization in rats. J Alzheimers Dis 2010, 21: 813–822.
Vicente E, Degerone D, Bohn L, Scornavaca F, Pimentel A, Leite MC, et al. Astroglial and cognitive effects of chronic cerebral hypoperfusion in the rat. Brain Res 2009, 1251: 204–212.
Davidson CM, Pappas BA, Stevens WD, Fortin T, Bennett SA. Chronic cerebral hypoperfusion: loss of pupillary reflex, visual impairment and retinal neurodegeneration. Brain Res 2000, 859: 96–103.
Crews D, McLachlan JA. Epigenetics, evolution, endocrine disruption, health, and disease. Endocrinology 2006, 147: S4–10.
de Carvalho CV, Payao SL, Smith MA. DNA methylation, ageing and ribosomal genes activity. Biogerontology 2000, 1: 357–361.
Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol Aging 2010, 31: 2025–2037.
Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev 2001, 15: 827–832.
Novakovic B, Wong NC, Sibson M, Ng HK, Morley R, Manuelpillai U, et al. DNA methylation-mediated down regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. J Biol Chem 2010, 285: 9583–9593.
Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol 2008, 8: 57–64.
Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci 2007, 31: 47–58.
McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci 2011, 31: 764–774.
Burns A, Iliffe S. Alzheimer’s disease. BMJ 2009, 338: b158.
Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiol Aging 2011, 32: 1161–1180.
Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med 2009, 46: 1241–1249.
West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci 1995, 6: 141–146.
Fuso A, Nicolia V, Pasqualato A, Fiorenza MT, Cavallaro RA, Scarpa S. Changes in Presenilin 1 gene methylation pattern in diet-induced B vitamin deficiency. Neurobiol Aging 2011, 32: 187–199.
Kwok JB. Role of epigenetics in Alzheimer’s and Parkinson’s disease. Epigenomics 2010, 2: 671–682
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, X., Sun, J. & Li, L. Chronic cerebrovascular hypoperfusion affects global DNA methylation and histone acetylation in rat brain. Neurosci. Bull. 29, 685–692 (2013). https://doi.org/10.1007/s12264-013-1345-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12264-013-1345-8