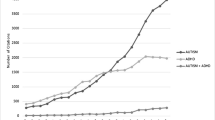
Abstract
In recent years, considerable progress has been made in understanding the genomic basis of autism spectrum disorder (ASD). Hundreds of variants have been proposed as predisposing to ASD, and the challenge now is to validate candidates and to understand how gene networks interact to produce ASD phenotypes. Genome-wide association and second-generation sequencing studies in particular have provided important indications about how to understand ASD on a molecular level, and we are beginning to see these experimental approaches translate into novel treatments and diagnostic tests. We review key studies in the field over the past five years and discuss some of the remaining technological and methodological challenges that remain.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Bailey A et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77.
Lauritsen MB, Pedersen CB, Mortensen PB. Effects of familial risk factors and place of birth on the risk of autism: a nationwide register-based study. J Child Psychol Psychiatry. 2005;46:963–71. doi:10.1111/j.1469-7610.2004.00391.x.
Sandin S et al. The familial risk of autism. JAMA. 2014;311:1770–7.
St Pourcain B. Variability in the common genetic architecture of social-communication spectrum phenotypes during childhood and adolescence. Mol Autism. 2014;5:18.
Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–95. doi:10.1016/j.tig.2007.06.003.
Hindorff L, MacArthur J, (2013). The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014 Jan;42(Database issue):D1001-6. doi:10.1093/nar/gkt1229.
Wang K et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–33. doi:10.1038/nature07999.
Ma D et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73:263–73. doi:10.1111/j.1469-1809.2009.00523.x.
St Pourcain B et al. Association between a high-risk autism locus on 5p14 and social communication spectrum phenotypes in the general population. Am J Psychiatry. 2010;167:1364–72. doi:10.1176/appi.ajp.2010.09121789.
Kerin T et al. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra140. doi:10.1126/scitranslmed.3003479. This study provides important functional validation of the 5p14. 1 locus as an important correlate of ASD susceptibility. Furthermore, it highlights a limitation in DNA sequencing, which would have failed to identify the MSNP1AS pseudogene.
Arking DE et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–4. doi:10.1016/j.ajhg.2007.09.015.
Alarcon M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70:60–71. doi:10.1086/338241.
Vernes SC et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–45. doi:10.1056/NEJMoa0802828.
Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–23. doi:10.1038/35097076.
Benayed R et al. Autism-associated haplotype affects the regulation of the homeobox gene, ENGRAILED 2. Biol Psychiatry. 2009;66:911–7. doi:10.1016/j.biopsych.2009.05.027.
Wang L et al. Association of the ENGRAILED 2 (EN2) gene with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:434–8. doi:10.1002/ajmg.b.30623.
Campbell DB et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–9. doi:10.1073/pnas.0605296103.
Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P. Genetic evidence implicating multiple genes in the MET receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res. 2008;1:159–68. doi:10.1002/aur.27.
Durand CM et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–7.
Moessner R et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–97. doi:10.1086/522590.
Peça J et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42.
McCauley J et al. Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid‐compulsive subset of autism. Am J Med Genet B Neuropsychiatr Genet. 2004;127:104–12.
Ro M et al. Association of the FGA and SLC6A4 Genes with autistic spectrum disorder in a Korean population. Neuropsychobiology. 2013;68:212–20.
Klei L et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3(1):9. doi:10.1186/2040-2392-3-9. This study used genome-wide complex trait analysis (GCTA) to confirm the importance of common variants as ASD risk factors, supporting an additive model of ASD where many variants of small effect combine to increase susceptibility.
Gaugler T et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–5.
Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17:764–72.
Cross-Disorder Group of the Psychiatric Genomics Consortium et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984-94. doi:10.1038/ng.2711.
Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–7. doi:10.1038/ng2084.
Abecasis GR et al. A map of human genome variation from population-scale sequencing. Nature;467: 1061-1073, doi:10.1038/nature09534 (2010).
Conrad DF et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–12. doi:10.1038/nature08516.
Pang AW et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11:R52. doi:10.1186/gb-2010-11-5-r52.
Sebat J et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi:10.1126/science.1138659.
Glessner JT, Connolly JJ, Hakonarson H. Rare genomic deletions and duplications and their role in neurodevelopmental disorders. Curr Top Behav Neurosci. 2012. doi:10.1007/7854_2011_179.
Bucan M et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi:10.1371/journal.pgen.1000536.
Kim HG et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi:10.1016/j.ajhg.2007.09.011.
Guilmatre A et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–56. doi:10.1001/archgenpsychiatry.2009.80.
Glessner JT et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi:10.1038/nature07953.
Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47:629–32. doi:10.1016/j.neuron.2005.07.008.
Missler M, Südhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol. 2012;4:a005694.
Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16 Spec No. 2:R168–73. doi:10.1093/hmg/ddm241.
Anney R et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–82. doi:10.1093/hmg/ddq307.
Hadley D et al. The impact of the metabotropic glutamate receptor and other gene family interaction networks on autism. Nat Commun. 2014;5:4074. doi:10.1038/ncomms5074. To find potentially druggable genetic targets, this study compared family interaction networks (GFINs) in 6742 patients ASDs and 12,544 neurologically normal controls, identifying enrichment of structural defects in the metabotropic glutamate receptor GFIN, as well as MXD-MYC-MAX and calmodulin 1 (CALM1) GFINs.
Povey S et al. The HUGO gene nomenclature committee (HGNC). Hum Genet. 2001;109:678–80.
Pinto D et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–72. doi:10.1038/nature09146.
Moreno-De-Luca D et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2012;18:1090–5.
Gilman SR et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi:10.1016/j.neuron.2011.05.021.
Sachidanandam R et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi:10.1038/35057149.
Sakai Y et al. Corticostriatal functional connectivity in non-medicated patients with obsessive-compulsive disorder. Eur Psychiatry. 2011;26:463–9.
Noh HJ et al. Network topologies and convergent aetiologies arising from deletions and duplications observed in individuals with autism. PLoS Genet. 2013;9:e1003523.
Li H et al. The association analysis of RELN and GRM8 genes with autistic spectrum disorder in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2008;147:194–200.
Serajee F, Zhong H, Nabi R, Huq AM. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J Med Genet. 2003;40:e42.
Cuscó I et al. Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet. 2009;18:1795–804.
Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–8.
Iossifov I et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi:10.1016/j.neuron.2012.04.009. Like several other studies in this review, Iossifov et al. use whole exome sequence to identify a range of mutations associated with ASD. Identification of the fragile X protein, FMRP, may have particularly important functional and translational consequences.
Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–6.
Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–7. doi:10.1016/j.tins.2004.04.009.
Jacquemont S et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 2011. doi:10.1126/scitranslmed.3001708.
Dolen G et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–62.
Richards RI et al. Fragile X syndrome: genetic localisation by linkage mapping of two microsatellite repeats FRAXAC1 and FRAXAC2 which immediately flank the fragile site. J Med Genet. 1991;28:818–23.
Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22:409–17.
Harris SW et al. Autism profiles of males with fragile X syndrome. Am J Ment Retard. 2008;113:427–38.
Curtis AR et al. X chromosome linkage studies in familial Rett syndrome. Hum Genet. 1993;90:551–5.
Ramocki MB et al. Autism and other neuropsychiatric symptoms are prevalent in individuals with MeCP2 duplication syndrome. Ann Neurol. 2009;66:771–82. doi:10.1002/ana.21715.
Muhle R, Trentacoste SV, Rapin I. The genetics of autism. Pediatrics. 2004;113:e472–86.
Carney RM et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–11.
Volkmar FR. Handbook of autism and pervasive developmental disorders. 3rd ed. New York: Wiley; 2005.
Li W et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–7. doi:10.1016/j.cub.2005.09.043.
Zafeiriou DI, Ververi A, Vargiami E. Childhood autism and associated comorbidities. Brain Dev. 2007;29:257–72. doi:10.1016/j.braindev.2006.09.003.
Schuurs-Hoeijmakers JH et al. Recurrent De Novo Mutations in <i> PACS1</i> cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am J Hum Genet. 2012;91:1122–7.
Rauch A et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82.
O'Roak BJ et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012. doi:10.1038/nature10989. The whole exome sequencing study showed that de novo germline mutations in coding regions are more prominent among males and found mutations in several existing ASD candidate genes.
Fischbach GD, Lord C. The Simons simplex collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi:10.1016/j.neuron.2010.10.006.
Hultman C, Sandin S, Levine S, Lichtenstein P, Reichenberg A. Advancing paternal age and risk of autism: new evidence from a population-based study and a meta-analysis of epidemiological studies. Mol Psychiatry. 2011;16:1203–12.
Malaspina D et al. Advancing paternal age and the risk of schizophrenia. Arch Gen Psychiatry. 2001;58:361–7.
McGrath JJ, et al. A Comprehensive Assessment of Parental Age and Psychiatric Disorders. JAMA psychiatry. 2014;71(3):301-9. doi: 10.1001/jamapsychiatry.2013.4081.
Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc Natl Acad Sci U S A. 2007;104:14801–6.
Neale BM et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012. doi:10.1038/nature11011. Whole exome sequencing study showed an association between de novo events and paternal age, as well as maternal age.
Sanders SJ et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012. doi:10.1038/nature10945. Whole exome sequencing study showing mutations in several ASD candidate genes. Of particular importance are two independent nonsense variants found to disrupt SCN2A.
Weiss LA et al. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry. 2003;8:186–94.
Jiang Y-h et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–63. Whole genome sequencing study if 32 families identifies mutations in several existing ASD candidate loci.
Kamiya K et al. A nonsense mutation of the sodium channel gene SCN2A in a patient with intractable epilepsy and mental decline. J Neurosci. 2004;24:2690–8.
Ogiwara I et al. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–53. doi:10.1212/WNL.0b013e3181b9cebc.
Phenome E, Consortium EK. De novo mutations in epileptic encephalopathies. Nature. 2013. doi:10.1038/nature12439.
de Ligt J et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Fromer M et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014. doi:10.1038/nature12929.
Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–96.
Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72.
Karakoc E et al. Detection of structural variants and indels within exome data. Nat Methods. 2012;9:176–8.
Glessner JT, Li J, Hakonarson H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013;41:e64. doi:10.1093/nar/gks1346.
Bölte S. Is autism curable? Dev Med Child Neurol. 2014. doi:10.1111/dmcn.12495.
Williams SC. Drugs targeting mGluR5 receptor offer 'fragile' hope for autism. Nat Med. 2012;18:840.
Pini G, et al. IGF1 as a potential treatment for Rett syndrome: safety assessment in six Rett patients. Autism research and treatment. 2012;679801. doi: 10.1155/2012/679801.
Ehninger D et al. Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat Med. 2008;14:843–8. doi:10.1038/nm1788.
Ehninger D, Silva AJ. Rapamycin for treating tuberous sclerosis and autism spectrum disorders. Trends Mol Med. 2011;17:78–87.
Silverman JL, Crawley JN. The promising trajectory of autism therapeutics discovery. Drug Discov Today. 2013. doi:10.1016/j.drudis.2013.12.007.
Silverman JL et al. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra151. doi:10.1126/scitranslmed.3003501.
Delorme R et al. Progress toward treatments for synaptic defects in autism. Nat Med. 2013;19:685–94.
Compliance with Ethics Guidelines
Conflict of Interest
John J. Connolly and Hakon Hakonarson declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This article is part of the Topical Collection on Autism Spectrum Disorders
Rights and permissions
About this article
Cite this article
Connolly, J.J., Hakonarson, H. Etiology of Autism Spectrum Disorder: A Genomics Perspective. Curr Psychiatry Rep 16, 501 (2014). https://doi.org/10.1007/s11920-014-0501-9
Published:
DOI: https://doi.org/10.1007/s11920-014-0501-9