Abstract
Attention-deficit/hyperactivity disorder, ADHD, is a common and highly heritable neuropsychiatric disorder that is seen in children and adults. Although heritability is estimated at around 76%, it has been hard to find genes underlying the disorder. ADHD is a multifactorial disorder, in which many genes, all with a small effect, are thought to cause the disorder in the presence of unfavorable environmental conditions. Whole genome linkage analyses have not yet lead to the identification of genes for ADHD, and results of candidate gene-based association studies have been able to explain only a tiny part of the genetic contribution to disease, either. A novel way of performing hypothesis-free analysis of the genome suitable for the identification of disease risk genes of considerably smaller effect is the genome-wide association study (GWAS). So far, five GWAS have been performed on the diagnosis of ADHD and related phenotypes. Four of these are based on a sample set of 958 parent–child trio’s collected as part of the International Multicentre ADHD Genetics (IMAGE) study and genotyped with funds from the Genetic Association Information Network (GAIN). The other is a pooled GWAS including adult patients with ADHD and controls. None of the papers reports any associations that are formally genome-wide significant after correction for multiple testing. There is also very limited overlap between studies, apart from an association with CDH13, which is reported in three of the studies. Little evidence supports an important role for the ‘classic’ ADHD genes, with possible exceptions for SLC9A9, NOS1 and CNR1. There is extensive overlap with findings from other psychiatric disorders. Though not genome-wide significant, findings from the individual studies converge to paint an interesting picture: whereas little evidence—as yet—points to a direct involvement of neurotransmitters (at least the classic dopaminergic, noradrenergic and serotonergic pathways) or regulators of neurotransmission, some suggestions are found for involvement of ‘new’ neurotransmission and cell–cell communication systems. A potential involvement of potassium channel subunits and regulators warrants further investigation. More basic processes also seem involved in ADHD, like cell division, adhesion (especially via cadherin and integrin systems), neuronal migration, and neuronal plasticity, as well as related transcription, cell polarity and extracellular matrix regulation, and cytoskeletal remodeling processes. In conclusion, the GWAS performed so far in ADHD, though far from conclusive, provide a first glimpse at genes for the disorder. Many more (much larger studies) will be needed. For this, collaboration between researchers as well as standardized protocols for phenotyping and DNA-collection will become increasingly important.
Similar content being viewed by others
Review
Attention-deficit/hyperactivity disorder (ADHD) is a common neuropsychiatric disorder. The prevalence of the disorder in children is estimated at 5.3%, estimates for adults vary from 1 to 4% (Faraone et al. 2000b; Kessler et al. 2006; Kooij et al. 2005; Polanczyk and Rohde 2007). A large number of studies, including family, adoption and twin studies suggest that ADHD is among the most heritable of neuropsychiatric disorders: ADHD was found to segregate in families with different studies finding risk increases of two to eightfold for parents and siblings of affected children (reviewed by Faraone and Doyle 2001). Adoption studies showed that the biological parents of hyperactive children carry a higher risk for ADHD compared to adoptive parents (Cantwell 1975; Morrison and Stewart 1973), and that first-degree adoptive relatives of probands with ADHD have a lower disease risk than the first-degree biological relatives of non-adopted ADHD probands (Sprich et al. 2000). More than 20 twin studies have been published in the last 32 years, most reporting estimates of heritability for ADHD between 60 and nearly 100%, with a mean of 76% (Faraone et al. 2005; Haberstick et al. 2008; Heiser et al. 2006; McLoughlin et al. 2007; Schultz et al. 2006).
The genetic architecture of ADHD is not currently clear. A polygenic transmission model seems likely (Faraone and Doyle 2001; Morrison and Stewart 1974), although some authors have suggested more dominant gene effects (Acosta et al. 2004; Deutsch et al. 1990; Maher et al. 1999; Morrison and Stewart 1974). Since it is also known that environmental factors play a role in ADHD etiology (for review see Banerjee et al. 2007), the disorder seems best described as being of multifactorial origin.
Identifying the individual genes contributing to the genetic variance of ADHD has proven difficult. Two approaches have been available for this type of research until recently: hypothesis-driven candidate gene association studies and hypothesis-free genome-wide linkage analysis. Although hundreds of candidate gene studies have been reported (see for example Bobb et al. 2005; Khan and Faraone 2006), only a handful of associations have been replicated across studies, although none of these achieved genome-wide significance (e.g. Faraone et al. 2005; Li et al. 2006). These studies have mainly concentrated on genes involved in neurotransmission, particularly in the catecholaminergic systems involved in the response to ADHD medications. Clearly, the hypothesis-driven approach has been limited by our restricted knowledge regarding mechanisms involved in ADHD pathogenesis. Genome-wide, hypothesis-free linkage analysis has been performed in ADHD using qualitative and quantitative definitions of the disease phenotype (Arcos-Burgos et al. 2004; Asherson et al. 2008; Bakker et al. 2003; Hebebrand et al. 2006; Ogdie et al. 2003; Ogdie et al. 2004; Ogdie et al. 2006; Romanos et al. 2008; Zhou et al. 2008a), and recently also using ADHD endophenotypes at the level of neuropsychological functioning (Doyle et al. 2008; Rommelse et al. 2008c). Although linkage designs have been highly successful in the identification of genes for monogenic diseases, this has not been the case for polygenic or multifactorial disorders. The linkage studies in ADHD have identified a number of genetic loci (potentially) harboring genes for ADHD, but very little overlap was observed between studies, so far. A recent meta-analysis of ADHD linkage studies only confirmed one locus, on chromosome 16 (Zhou et al. 2008c).
Generally, the results of both candidate gene-based association studies and genome-wide linkage studies have not identified major genes for ADHD. In candidate gene studies, individual gene variants have only shown small effects, rarely reaching an odds ratio of 1.3 (e.g. Faraone et al. 2005). This strongly limits the power of genetic linkage designs in ADHD research, as these designs are suited for the identification of individual genetic variants that explain at least 10% of the genetic variance of a trait or disease (Risch and Merikangas 1996). A situation like this calls for a new analytic approach, one which combines the power to detect genetic variants of small effect size, like the association studies, with the possibility to perform hypothesis-free analyses of the entire genome. This has recently become feasible using the genome-wide association study (GWAS) methodology. In GWAS, 100,000 to more than 1,000,000 genetic variants (in this case single nucleotide polymorphisms, SNPs), distributed across the entire genome are genotyped using a microarray platform and (individually) tested for association with a trait or disorder. The selection of SNPs is mostly based on the distribution of linkage disequilibrium (LD) across the genome, as identified by the HapMap project (Frazer et al. 2007; The International HapMap Consortium 2005). This selection is sufficient to obtain information about most of the frequent genetic variation in the genome (Manolio et al. 2008; Pearson and Manolio 2008), especially for the newer platforms, which include around 1,000,000 or more SNPs. In the short time of their existence, GWAS have already led to the identification of more than 100 genetic variants significantly associated with about 40 different traits and diseases, including a number of psychiatric ones (e.g. Ferreira et al. 2008; O’Donovan et al. 2008; O’Donovan et al. 2009; Sklar et al. 2008). Although highly successful, design-wise (for an overview of GWAS approaches see Neale and Purcell 2008), the GWAS method has not been exploited to the fullest, given its high financial costs (sample sizes of several thousands to ten-thousands may be needed, see for a recent discussion of this matter Burton et al. 2008). Major progress in the field came through a number of GWAS initiatives funded by government and industry, like the Welcome Trust Case Control Consortium (WTCCC; 2007) and the Genetic Association Information Network (GAIN; Manolio et al. 2007). Among the 13 diseases that were analyzed in the first rounds of these two initiatives, the WTCCC included 1 and GAIN included 4 psychiatric disorders.
GAIN has also been the setting for the first GWAS in ADHD, which was carried out in 958 Caucasian case–parent-trios collected as part of the International Multicentre ADHD Genetics (IMAGE) study in children (Brookes et al. 2006; Kuntsi et al. 2006). Using the genotyping data from this study, Neale et al. (2008a) reported a classic transmission disequilibrium analysis (TDT) of a categorically defined ADHD phenotype. In addition, Lasky-Su et al. (2008b) performed an analysis using quantitative measures of ADHD symptoms. In the meantime, a second set of genome-wide data has been reported by Lesch and coworkers. These researchers used pools of DNA from 343 ADHD-affected adults and 304 controls for their genome-wide association analysis of a categorical ADHD phenotype (Lesch et al. 2008). Following the first analyses, the GAIN/IMAGE ADHD dataset has since been used for a number of exploratory studies using additional phenotypes and designs, like the age of onset of ADHD symptoms (Lasky-Su et al. 2008a), conduct problems (Anney et al. 2008b), and even a genome-wide gene–environment interaction (G × E) study evaluating genetic variants moderating the effects of maternal expressed emotion on ADHD symptoms (and conduct problems; Sonuga-Barke et al. 2008). In the following paragraphs, we will discuss most of these studies and their most important findings. The two studies exploring conduct problems in the GAIN/IMAGE dataset (Anney et al. 2008b; Sonuga-Barke et al. 2008) are beyond the scope of this review.
As mentioned above, most of the GWAS data so far are derived from one sample of child–parent triads collected as part of the IMAGE study. IMAGE is a consortium of researchers from seven European countries and Israel. IMAGE has ascertained more than 1,400 families through a (preferentially combined subtype) ADHD-affected proband (Brookes et al. 2006; Kuntsi et al. 2006). The probands were required to be in the age range from 5 to 17 years. All families were of European Caucasian descent. In all families, both parents and at least one sibling, the latter not selected for phenotype, were required to take part in the study. IMAGE participants were extensively phenotyped for ADHD and comorbid disorders, using the Parental Account of Childhood Symptoms (PACS) (Taylor et al. 1986), administered by centrally trained investigators. In addition, rating scale measures were used, including the Long Version of Conners’ Parent and Teacher Rating Scales, as well as the parent and teacher version of the Strength and Difficulties Questionnaires (SDQ) (Conners 2003). A standardized algorithm was applied to PACS to derive each of the 18 DSM-IV ADHD items, and, taking into account a number of Conners items, a clinical diagnosis was made. Exclusion criteria included a low IQ (<70), autism, epilepsy, and brain or genetic disorders known to mimic ADHD symptoms. In GAIN, 958 family triads from IMAGE, including the proband and his/her parents were analyzed (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000016.v1.p1). DNA was isolated from blood by Rutgers Cell and DNA Repository (http://www.rucdr.org/). The samples were analyzed by Perlegen Sciences on a microarray specially designed for GAIN. The array investigates approximately 600,000 SNPs able to capture information of close to 100% of the common genetic variation in Caucasians (Neale et al. 2008a). Data cleaning of the genotyping results was performed through the National Centre for Biotechnology Information (NCBI), leaving 438,784 SNPs for analysis.
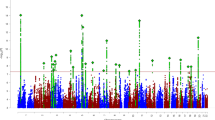
The first paper on the GAIN/IMAGE ADHD genome-wide dataset was based on the 909 complete family triads, for which full genotyping information was available (Neale et al. 2008a). Of these families, 790 contained male probands, 119 female probands. A total of 845 probands had a combined subtype ADHD diagnosis, the average number of symptoms was 16.1. The TDT design was chosen for this study which contained families recruited from all over Europe and from Israel, based on its robustness against population stratification. Earlier research by Neale et al. (2008b) had shown that differences indeed existed between the different genetic backgrounds of the IMAGE participants, especially with the Israeli families of European Caucasian descent. In addition, the family controlled design showed more stability in the presence of non-random genotyping error in GAIN (Anney et al. 2008a). No genome-wide significant results were observed (with the highest P values at 7.45E-06), although a power analysis presented in the paper suggests that the size of the dataset would have been sufficient to detect at least one genome-wide significant (P value 5.0E-08) finding for a genetic variant with a minor allele frequency (MAF) of 0.2 causing an odds ratio of 1.3. A table in the Neale et al. paper presents the highest-ranking 25 SNPs (based on P value) (see also Table 1 in this review).
In a TDT design, the number of observed transmissions of a given allele of a SNP is compared to the expected frequency under the assumption of no association (Spielman and Ewens 1996). Performing the TDT, the authors noted a highly significant imbalance in the number of major and minor allele over-transmissions. Given the large numbers of SNPs analyzed, these numbers would have been expected to be similar. Removing additional SNPs that had failed quality control in one of the other two GWAS based on the Perlegen microarray improved this imbalance partly, suggesting that genotyping error or missingness is (at least partly) responsible for this effect. Therefore, the authors repeated their analysis including a correction factor. Comparison of the presented tables listing the top-25 SNPs before and after correction shows that this resulted in the loss of half of the listed SNPs and a general downward correction of P values. Although, as the authors state, it might be (too) early to explore the biological relevance of the strongest observed associations, it is a fact that the list contains a number of very interesting candidate genes for ADHD (Table 1), most notably those genes that also show association in the GWAS of quantitative ADHD-related phenotypes (Lasky-Su et al. 2008b), as indicated in Table 1. Among these is a SNP close to CNR1, which encodes the cannabinoid receptor 1, a gene that falls into a linkage region for ADHD from a primary analysis (Ogdie et al. 2004) and shows suggestive linkage in meta-analysis (Zhou et al. 2008c). CNR1 was also found associated with the disorder in candidate gene-based association studies (Lu et al. 2008; Ponce et al. 2003). Furthermore, association of CNR1 with alcohol and drug abuse and dependence has been observed (Zuo et al. 2007). Also of interest are the findings for the especially brain-expressed cytoskeleton-organizer DCLK1 and the extracellular matrix component SPOCK3, as well as the two potassium-channel regulators KCNIP1 and KCNIP4 (Table 1).
In an attempt to maximize the power of the GAIN/IMAGE family-based GWAS, Lasky-Su et al. (2008b) published a study focusing on quantitative ADHD phenotypes using the Family-based Association Test (FBAT) suite of programs. Based on the 18 DSM-IV symptoms of ADHD the authors constructed 3 quantitative phenotypes using (1) the number of hyperactive–impulsive symptoms, (2) the number of inattentive symptoms, and (3) the total number of symptoms. These phenotypes were each tested under three different genetic models (additive, dominant, recessive). Furthermore, three additional quantitative traits were constructed based on the Conners ADHD Rating Scales and the PACS using FBAT-PC methodology (Lange et al. 2004c). This approach constructs a slightly different phenotype for every SNP in order to maximize the heritability at this SNP. Association testing was subsequently limited to the set of ten SNPs per genotype with the highest power to detect association derived from a screening algorithm in the program PBAT (Lange et al. 2004b; van Steen and Lange 2005). In total, this study performed 18 GWAS, increasing the multiple testing burden; correction for multiple testing was limited to the single GWAS.
In addition to increasing power, using symptoms or rating scales as a basis for phenotype selection had the advantage that hyperactive–impulsive and inattentive symptoms could be assessed independently in the combined subtype ADHD of the GAIN/IMAGE participants. However, a prerequisite for this quantitative approach to be valid is that ADHD is a disorder at the extreme of a continuum observed in the population. Evidence suggesting that this is indeed true comes from twin studies showing strong heritability of quantitative measures of ADHD (Lasky-Su et al. 2008b, and references herein) as well as from a proband–sibling comparison in IMAGE (Chen et al. 2008; Thapar et al. 2006). Starting from the cleaned GAIN dataset, Lasky-Su restricted the analysis to 429,784 autosomal SNPs (as FBAT cannot handle X-linked markers) and 909 complete families. 87% of the probands were male, and the mean age of probands was 10.88 years. The average total number of symptoms was 16.11, the number of hyperactive–impulsive symptoms was 8.11 and the number of inattentive ones was 7.98.
Two genome-wide significant findings (corrected for single analysis) emerged from the formal tests, one for rs6565113 in an intron of CDH13 associated under an additive genetic model with the FBAT-PC-based phenotype derived from all ADHD symptoms, one for rs552655 in an intron of GFOD1 associated with the FBAT-PC phenotype for inattentive symptoms (dominant model). The CDH13 finding is especially interesting, since a SNP near this gene also is part of the top-25 in the GWAS by Neale et al. (2008a, b), though the two studies are not entirely independent, of course, and the gene is also found among the top-findings from the ADHD GWAS by Lesch et al. (2008, see below) and falls into the only significant ADHD linkage region identified in the recent meta-analysis (Zhou et al. 2008c). Furthermore, association with SNPs in CDH13 is one of the most consistent findings in genome-wide studies on a wide variety of phenotypes related to drug abuse and dependence (Uhl et al. 2008a, b), although there is no clear indication, which region of the gene is most important. Possibly, different regions or risk factors might cause different phenotypes. CDH13 codes for cadherin 13 (or T-cadherin), a member of a family of cell–cell adhesion proteins (Patel et al. 2003), in addition to being a regulator of neural cell growth. CDH13 shows a brain expression profile consistent with a role in ADHD, being expressed in regions showing volumetric reductions in patients with this disorder (Takeuchi et al. 2000; Takeuchi and Ohtsuki 2001; Valera et al. 2007). Very little is known about the second associated gene, GFOD1 and its product, glucose–fructose oxidoreductase-domain containing 1, except that it is expressed in brain and possibly plays a role in electron transport (Lasky-Su et al. 2008b).
In addition to the formal testing of ten SNPs for each phenotype Lasky-Su and coworkers also report a more exploratory analysis of all SNPs in the dataset. There were 58 association findings with P values smaller than 10−5, of which 46 SNPs were unique. Some SNPs showed association under more than one genetic model and/or across phenotypes. Although none of the SNPs listed here are genome-wide significantly associated with ADHD, a high percentage of the SNPs is present in brain-expressed genes and/or in linkage regions for ADHD and other psychiatric disorders (see Table 2). A particularly interesting finding from this set includes the SNP in NOS1, encoding the neuronal form of nitric oxide synthase. NO acts as the second messenger of the N-methyl-d-aspartate receptor and interacts with both the dopaminergic as well as the serotonergic system in the human brain. NOS1 has been associated with impulsive and aggressive behavior and ADHD (Reif et al. 2009). Association has also been noted with Alzheimer’s disease (Galimberti et al. 2008) and schizophrenia, as well as with related neuropsychological performance (Reif et al. 2006).
Lasky-Su and colleagues also reported P values for a selection of predefined (classic) ADHD candidate genes (Brookes et al. 2006). Most significant findings with P values in the range of 10−5 were observed for SLC9A9. This gene (which was with a total of 181 SNPs the largest gene analyzed) also contained the largest number of associations in terms of SNPs and phenotypes. Other findings at P values of 10−4 were for DDC and SNAP25. The SLC6A1, ADRB2, HTR1E, ADRA1A, DBH, BDNF, DRD2, TPH2, HTR2A, SLC6A2, PER1, CHRNA4, COMT and SYT1 genes showed association at 10−3. SLC9A9 encodes a sodium/hydrogen exchanger and may be a particularly viable candidate gene for ADHD. It was originally reported disrupted in a patient with ADHD (de Silva et al. 2003) and also shows mutations in patients with autism (Morrow et al. 2008) as well as having been found in multiple GWAS for addiction-related disorders (Uhl et al. 2008a, b).
A GWAS not based on the GAIN/IMAGE sample was published by Lesch et al. (2008). In their study they analyzed 343 German patients with persistent ADHD (mean age 32.9 years with a range from 18 to 65, 54.5% male, 61.5% combined subtype, 31.2% inattentive subtype, 7.3% hyperactive–impulsive subtype) and 304 controls (mean age 32.7, 51.3% male). Patients were diagnosed using the Structured Clinical Interview of DSM-IV axis 1 disorders (SCID-I; Jacob et al. 2007), supported by ratings from the Wender Utah Rating Scale and an ADHD diagnostic checklist applying DSM-IV criteria categorically and dimensionally (ADHD-DC) (Rosler et al. 2004). Comorbidity along the entire spectrum was analyzed using SCID-I and SCID-II, the latter assessing axis II disorders. Lifetime comorbidity with major depression was 44.7%, with substance use disorders it was 44.1% (of which 32.0% for cannabis use, 40.6% for alcohol use). Of the 304 controls only 54 were screened for psychiatric disorders prior to inclusion. Genotyping was on the Affymetrix GeneChip Human Mapping 500K sets. Importantly, instead of genotyping individual samples, pools were constructed of the 343 cases and 304 controls, respectively. For each pool nine technical replicates were analyzed. After data cleaning the authors ended up with allele frequencies for 504,219 SNPs, of which the autosomal SNPs were ranked based on the mean of rankings performed using ANOVA, rank sums and silhouette scores. Whereas the other ADHD GWAS reports relied strictly on statistical rankings to describe their results, Lesch and coworkers used additional criteria for further prioritization of SNPs for follow up, one being localization within or near genes (±100 kb flanking region), a second being presence in clusters of at least three SNPs located in regions of genes, a third being the comparison with a family-based association study performed in a dataset from a recently published linkage paper by the same group, or being present in a suggestive linkage region from this analysis in adult ADHD patients (Romanos et al. 2008). In addition, expression in brain, as well as earlier findings suggesting involvement in complex traits and neuropsychiatric disorders (especially substance use disorders) were taken into consideration.
Although none of the findings were genome-wide significant, interesting candidate genes for ADHD are reported in this paper, too. Of the 80 top-ranked SNP findings, 30 fall into genes, of which nearly all are known to be expressed in brain (see Table 3). A second list, containing the top-10 of those genes with 3 or more SNPs among the 1,000 highest-ranking findings contains the genes KALRN (15 SNPs), ZNF354C (15 SNPs), WRNIP1 (14 SNPs), GRB10 (10 SNPs), DPP6 (7 SNPs), ARHGAP22 (11 SNPs), RAB38 (11 SNPs), FAT3 (8 SNPs), DA259379 (13 SNPs), NT5DC3 (20 SNPs). A third list summarizes all findings that show overlap with the linkage analysis in adult ADHD patients published earlier by the same group (Romanos et al. 2008), and in some instances also a family-based GWAS based on the 50 K linkage dataset (Table 3 in Lesch et al. 2008). Of the 30 genes listed there, 8 genes (CTNNA2, MOBP, MAP1B, REEP5, ASTN2, ATP2C2, CDH13 and ITGAE) overlap with the list of the 80 top-ranked SNP findings, as indicated in Table 3. The authors highlight the findings for ASTN2, CSMD2, ITGA11, CTNNA2 and CDH13 in their discussion. All of these genes have earlier shown association with substance use disorders and related phenotypes, which is interesting in the light of the frequent comorbidity with such disorders in (adult) ADHD (44.1% in the current study). The five genes encode proteins involved in cell adhesion and/or cell–cell communication in development and maturation of the brain. Also the UNC5B (Table 3) and KALRN genes fit into this category. The latter encodes a protein present in the postsynaptic density which regulates dendritic spine development and is important for the neuronal plasticity underlying memory and learning (Lesch et al. 2008; Penzes and Jones 2008).
Comparing this list with the findings from the other two GWAS, very little overlap is seen (which might be expected, given power limitations of the studies, see below), except for one finding on chromosome 16, which falls into CDH13. As indicated above, this gene was also among the top-findings from both GAIN/IMAGE GWAS studies, as well as many genome-wide studies of addiction related phenotypes, recently reviewed by Uhl et al. (2008a). An additional similarity to be noted is for two TLL genes, TLL1 found among the top-ranks of the analysis based in the categorical ADHD phenotype and showing nominal association in the quantitative analyses by Lasky-Su et al. (see Table V in Lasky-Su et al. 2008b), TLL2 present among the top-30 SNPs in gene regions in the analysis by Lesch et al. Both genes encode metalloproteases that cleave collagen and are expressed in multiple tissues including brain. A specific role in brain development or functioning has not been reported for either of these genes, though. Additional genes modulating the extracellular matrix are found in all three studies, including HAS3 (Lasky-Su et al. 2008b), SPOCK3 (Neale et al. 2008a), MAN2A2, GPC6 and MMP24 (Lesch et al. 2008). A third interesting overlap is for genes related to potassium-channel function, like the KCNC1 gene reported by Lesch et al. (2008), the KCNIP1 and KCNIP4 genes observed by Neale et al. (2008a)) and the DPP10 finding for total ADHD symptom count (Lasky-Su et al. 2008b).
An exploratory analysis based on the GAIN/IMAGE sample made use of the age of onset of hyperactive–impulsive and inattentive symptoms, as assessed with a question from the PACS interview, each, as an informative phenotype for a GWAS (Lasky-Su et al. 2008a). Earlier work of the authors had suggested that environmental exposure outside the womb were less likely to contribute significantly to the development of ADHD with a very early age of onset (Lasky-Su et al. 2007). The number of children included in the analysis was 930 with a mean age of 10.86 years, 810 of them being males. The average age at onset of ADHD symptoms was 2.67 years; for the hyperactive–impulsive symptoms this age was 2.77, for the inattentive symptoms it was 4.31 years. The study investigated 429,784 autosomal SNPs using the FBAT-logrank algorithm (Lange et al. 2004a) to identify genetic variants that could predispose children to an earlier age of onset of disease symptoms. Additive, dominant and recessive genetic models were tested. A total of 16 SNPs reached P values for association <10−5 including 14 unique findings (Table 4). There is no overlap with the results from the other GWAS. Although potentially due to chance it is remarkable to see that this list contains a number of genes that are involved in the response of the body to environmental exposures and/or are regulated by the environment, like the inflammation related ADAMTS2, the stress regulator MAP3K7 or the vitamin A-responsive NAV2 gene.
An interesting new approach in GWAS is represented by the exploratory study by Sonuga-Barke et al. (2008). The author and his colleagues evaluated gene-environment interactions (G × E) in ADHD in a genome-wide analysis. Specifically, they investigated the effects of maternal warmth and criticism on ADHD severity in the GAIN/IMAGE dataset (also, a G × E analysis for conduct disorder symptoms is reported in the publication, but is not reviewed here). ADHD symptoms were derived from the PACS interview in IMAGE, maternal expressed emotion was assessed using the Camberwell Family Interview. Maternal warmth and criticism were coded, separately; warmth was assessed by tone of voice, spontaneity, sympathy and/or empathy toward the child. Criticism was coded from statements that criticized or found fault with the child based on the tone of voice and critical phrases (Sonuga-Barke et al. 2008). Statistical analysis made use of the FBAT-Interaction methodology (Vansteelandt et al. 2008). In total, 430,060 autosomal SNPs were included in the analysis, as were 909 patients (87% males) from complete trios. Mean age of the participants was 10.88 years, the average number of ADHD symptoms was 16.1, the mean overall measure for mother’s warmth was 1.43 (range from 0 to 3) and for maternal criticism it was 1.74 (range from 0 to 4). Additive, dominant and recessive genetic models were tested for the phenotype and the two environmental factors.
The analysis did not produce any genome-wide significant findings, the highest P value reaching 9.08 × 10−7. Nineteen SNPs showed interaction association P values below 10−5 in the presence of main genetic effects (at P ≤ 10−5), of which five showed association under two genetic models (Table 5). Only two findings were related to maternal criticism. Most of the genes interacting with maternal warmth clustered in three distinct regions on chromosomes 6, 11 and 13. The regions on chromosomes 6 and 11 fall into genes, KIF6 and PIWIL4, respectively. The region of 58 kb on chromosome 13 includes 8 association findings for 4 unique SNPs, and although it does not contain genes, it shows extensive LD extending proximally to the SLC46A3 gene, the function of which is unknown. Interestingly, the KIF6 gene has repeatedly been shown to respond to environmental stimuli (Iakoubova et al. 2008b; Iakoubova et al. 2008a; Shiffman et al. 2008). In the referenced studies, a coding variant in KIF6 altered the response to statins in cardiovascular disease. The gene’s role in moderating the influence of mother’s warmth on ADHD severity will have to be investigated in further detail. An additional six interaction associations at P values of 10−4 were observed in the absence of genetic main effects (Table V in Sonuga-Barke et al. 2008). Interestingly, all but one finding were observed under a recessive genetic model, which strongly limited the number of informative families. Of these associations, only one was for mother’s warmth. This finding falls into the BCAS1 gene on chromosome 20, which is a potential oncogene with high expression in brain. Of the five association findings interacting with mother’s criticism, especially a finding on chromosome 2 is interesting, as it falls into the MTA3 gene, which has two additional SNPs associated with ADHD in the GWAS for quantitative phenotypes (Lasky-Su et al. 2008b). The authors also tested potential G × E effects for the list of ADHD candidate genes defined earlier (Brookes et al. 2006). Interaction association P values are most significant for DDC (mother’s warmth and criticism), HTR2A and SLC9A9 (warmth) at 10−4.
The interpretation of the analytical strategy used in this study is difficult. First of all, the direction of causation is not clear: it could be that maternal expressed emotion influences ADHD severity in the child, on the other hand, a child with severe ADHD might also elicit more pronounced criticism and less warmth from parents. Furthermore, explaining the biological effects of the environmental moderators of ADHD severity is not straightforward. The authors discuss epigenetic effects (like DNA methylation) as possible mechanisms. This might indeed be supported by at least one of their findings: PIWIL4 plays a role in chromatin-modification, another epigenetic regulator of gene expression (Szyf et al. 2007, 2008).
Taking together all results from the GWAS performed in ADHD, the following is to be remarked: none of the findings so far show genome-wide significant association with ADHD according to the thresholds currently handled (Dudbridge and Gusnanto 2008; Pe’er et al. 2008; van Steen and Lange 2005). Nevertheless, the most high-ranking findings from the different studies contain interesting new candidates for further study. Little overlap is observed between studies, except for CDH13, which is found in three studies (Lasky-Su et al. 2008b; Lesch et al. 2008; Neale et al. 2008a). None of the ‘classic’ candidate genes for ADHD are found among the top-findings of either study, although two more recently reported genes, NOS1 and CNR1, are observed at least once. In the candidate gene analyses reported for the GAIN/IMAGE sample, most evidence is repeatedly found for involvement of SLC9A9 in ADHD. For all studies discussed here, overlap with results from linkage studies reported in ADHD and linkage as well as GWAS in other psychiatric disorders is multiple.
Looking at the processes potentially involved in the etiology of ADHD, little evidence (as yet) points to a direct involvement of neurotransmitters (at least the classic dopaminergic, noradrenergic and serotonergic pathways) or regulators of neurotransmission (except for DNM1). Some suggestions are found for involvement of ‘new’ neurotransmission and cell–cell communication systems, though, like glutamate, vasopressin, and TAFA proteins. Also, the potential involvement of potassium channel subunits and regulators definitely warrants further investigation. However, more basic processes also seem involved, like cell division, adhesion (especially via cadherin and integrin systems), neuronal migration, and neuronal plasticity, as well as related transcription, cell polarity and extracellular matrix regulation, and cytoskeletal remodeling processes. Our conclusion is consistent with and extends the conclusion drawn by Lesch and coworkers based on the findings from their GWAS, in which they suggest that neuronal spine formation and plasticity may underlie the pathophysiology of ADHD. Finding inflammatory genes (IL16 and ADAMTS2) is somewhat surprising, although inflammation is already known to be involved in different psychiatric disorders, like major depression and Alzheimer’s disease (Anisman 2009; Bazar et al. 2006; Garcia-Bueno et al. 2008; Serretti et al. 2007). The analysis of age of onset of ADHD revealed a number of environmentally regulated genes (Lasky-Su et al. 2008a), which may suggest a particularly important influence of the environment in this aspect of the disorder. However, all of these biological hypotheses must be tempered by the limited knowledge of the genetic architecture of ADHD at this point. Currently, we do not have sufficient information to draw strong conclusions about the relative impact of the biological pathways and genetic influences, aside from likely knowing that the genetic effects are modest in size.
In the last 2 years, much progress has been made in identifying genes for psychiatric disorders through GWAS. From the studies so far it has become apparent that early power estimates have been far too optimistic and at least several thousands of samples will be needed for the design of an optimal study (Burton et al. 2008). Genetic heterogeneity appears even more extensive than previously expected, with relative risks due to single genetic variants in the range of 1.05–1.1 or even smaller (Ferreira et al. 2008; Neale et al. 2008a; O’Donovan et al. 2008; Sklar et al. 2008). ADHD is no exception to this, and so all of the ADHD GWAS performed to date are highly underpowered. In addition, Lesch and colleagues used a pooling design, which reduces power by about 70% and puts limitations to the genetic models that can be investigated (Lesch et al., 2008). Furthermore, using controls unscreened for the presence of psychiatric disorders can be expected to reduce power, though the effect of this is modest, given that the prevalence is at most 5% (McCarthy et al. 2008). The X-chromosome was not analyzed in most of the studies, due to a limitation of the FBAT-program. As a clear difference in prevalence of ADHD exists between genders, this might still be an interesting target to look at.
An important issue in psychiatric disorders in general and ADHD in particular is phenotypic heterogeneity. First, psychiatric diagnosis must rely on clinical symptoms that, without external validators, cannot be expected to reflect biologically relevant concepts that are closely linked to genes (Kuntsi et al. 2006; Thapar et al. 2006). Although the instruments used to define psychiatric GWAS phenotypes are generally chosen for high reliability, they have no proven biological validity and will therefore (at best) result in a more ‘noisy’ phenotype definition than most biological tests (Craddock et al. 2008). More importantly, however, the classical nosological approaches identify phenotypically distinct diseases, such as ADHD, substance abuse, affective disorder and schizophrenia, though these exhibit considerable overlap in symptomatology, clinical course, and long-term outcome. ADHD is hardly ever the only disorder a patient suffers from, comorbidity is the rule rather than the exception. For example, reported frequencies of depression and substance use disorders were also high in the GWAS sample used by Lesch et al. (2008). These factors severely hamper the power of genetic association studies and might explain why psychiatric genetics has had an extremely modest track record in pinpointing and characterizing susceptibility loci, even using the GWAS approaches.
That said, what are the perspectives for genome-wide association studies in ADHD? The results of the first studies—though most not genome-wide significant—are already very exciting and change the way in which we think about genetic factors involved in ADHD etiology. For now, GWAS holds the greatest promise for understanding the genetic architecture of ADHD! However, we need to improve the design of these studies by increasing sample size for more power and by further improving/extending phenotypic assessment to better reflect and partition the phenotypic complexity of the disorder. The former will only become possible through collaboration. As a field, ADHD has the good fortune of having several thousand samples collected world-wide, with collaborative efforts supported by the National Institute of Mental Health. Investigators with ADHD GWAS datasets are also participating in the Psychiatric GWAS Consortium (PGC), a confederation of 101 scientists from 11 countries and 48 institutions having GWAS datasets on ADHD, autism, bipolar disorder, schizophrenia and major depression (Psychiatric GWAS Consortium Steering Committee 2009). The PGC investigators have agreed to participate in coordinated mega-analyses both within and across disorders. Within 1 year, it is to be expected that a collaborative GWAS with more than 4,000 cases can be carried out via the PGC for ADHD. It is this spirit of collaboration that we believe will be needed to achieve the breakthroughs in genetic research that will ultimately help us discover new pathophysiological pathways and targets for treatment.
Improving GWAS design through extension of phenotypic assessment could be done by taking characteristics into consideration that we already know or suspect to play a role in genetic susceptibility. Given the limits of the nosological systems, subdividing ADHD according to subtype will probably not be sufficient, as there is already evidence of shared genetic factors between subtypes (Faraone et al. 2000a; Thapar et al. 2006). Taking into account comorbidity might deal with some of the limits of the phenotypes, as a number of studies have suggested that ADHD plus a specific comorbidity may constitute a distinct subtype of ADHD (Faraone et al. 2000b; Faraone 2000; Faraone and Doyle 2001; Fliers et al. 2009; Rasmussen et al. 2004; Smalley et al. 2000; Willcutt et al. 2000). A recent study in IMAGE can be used as an example that this affects association findings: Zhou et al. (2008b) showed that SLC6A3/DAT1 was only associated with ADHD in the absence of conduct disorder. Another example is gender; given the difference between sexes, certain genetic risk factors can be expected to affect males and females differently. That this affects association studies has already been shown in ADHD (Biederman et al. 2008; Guimaraes et al. 2007; Rommelse et al. 2008b) as well as other (related) psychiatric disorders (Craig et al. 2004; Diamond 2007; Haefner et al. 2008; Kendler 1998; Lang et al. 2008; Okamura et al. 2007; Rucci et al. 2009). An additional potentially important factor is age at diagnosis. It is well established that age influences cognitive performance, with children performing worse in neuropsychological tests than adolescents and adults (Addamo et al. 2007) on tasks that are relevant to ADHD (Rommelse et al. 2008a). We and others have presented evidence that age can modulate the association of SLC6A3/DAT1 with ADHD, with different haplotypes of the gene being associated with ADHD in childhood and adulthood (Barkley et al. 2006; Elia and Devoto 2007; Franke et al. 2008).
A more radical way of dealing with the shortcomings of the nosological system is to look deeper into the possibilities of using (ADHD) endophenotypes for GWAS. Endophenotypes, or intermediate phenotypes, represent heritable phenotypic constructs that are, presumably, more directly affected by genes than clinical symptoms or disease categories (Gottesman and Gould 2003; Szatmari et al. 2007a; Walters and Owen 2007). This should result in a stronger association between causal genes and the endophenotype than the clinical disease phenotype. The success of an endophenotype strategy requires either a higher heritability of the endophenotype compared to the disease phenotype (which is generally not generally observed, Szatmari et al. 2007a) or a reduced complexity of the genetic architecture of the endophenotype due to the involvement of less genes (see Fig. 1). Endophenotypes for ADHD have been proposed at different levels, e.g. those based on neuropsychological performance (Doyle et al. 2005; Rommelse 2008) and those based on neuroimaging (Castellanos and Tannock 2002), i.e. brain activity and structure. Recent genome-wide linkage studies of neuropsychological endophenotypes for ADHD indeed suggest that these measure offer increased power for finding loci compared to clinical phenotypes (Doyle et al. 2008; Rommelse et al. 2008c). A pooled GWAS on memory performance also suggests this (Papassotiropoulos et al. 2006). ‘Imaging genetics’ studies would be thought to be particularly powerful, requiring even smaller sample sizes (Dreher et al. 2008; van Haren et al. 2008). Studies using such ADHD endophenotypes for GWAS are currently ongoing.
Simplified schematic representation of the endophenotype concept in psychiatric genetics. Many genes (depicted in an oversimplified way as gene A to I) are involved in causing a disease symptom and the categorical disease phenotype. A reduced number of genes is involved in disease related endophenotypes, like ‘Function of a brain unit’ (Level 1), as assessed through performance on neuropsychological tests; even less genes play a role if an endophenotype at levels 2 or 3 is used for genetic association studies. On level 2, endophenotypes measured through (structural and functional) neuroimaging are shown, on level 3 cellular behavior, assessed through e.g. assays measuring migration, apoptosis or cell division rates are shown
An additional aspect to be taken into consideration in GWAS is the potential role of G × E and gene–gene (G × G) interactions in the etiology of ADHD. As the exploratory analysis by Sonuga-Barke et al. (2008) shows, this type of analysis will help to further delineate mechanisms moderating disease severity and those leading to disease.
A last aspect to be discussed relates to the genetic models currently used in ADHD research. Up to now, we work under the assumption that the ‘common disease, common variant’ model will hold for ADHD. This model suggests that ADHD is caused by multiple variants that are common in the population and each contribute a small amount of disease risk. However, experience from GWAS in other multifactorial disorders now challenges this view and postulates the existence of rare genetic variants contributing to disease, that are not captured by GWAS (see also McCarthy et al. 2008 for a more elaborate discussion of this subject). This case of the ‘missing heritability’ might imply that private or low-frequency variants of large effect size exist and actually explain most of the heritability of a multifactorial disorder in individual patients. Copy number variants (CNVs) might contribute a part of this puzzle. CNVs include insertions, deletions and duplications and encompass relatively large genomic segments of 1 kb to several Mb in size (Redon et al. 2006; Sebat et al. 2004). Although they are a frequent class of polymorphism in the human genome, most CNVs are rare. Many reports indeed now suggest involvement of CNVs in psychiatric disorders (Lachman et al. 2007; Marshall et al. 2008; Sebat et al. 2007; Stefansson et al. 2002, 2008; Szatmari et al. 2007b; Ullmann et al. 2007; Vrijenhoek et al. 2008; Walsh et al. 2008; Weiss et al. 2008). The role of CNVs in ADHD has not been studied, yet.
In conclusion, the GWAS performed so far in ADHD, though far from conclusive, provide a first glimpse at genes for the disorder. Many more (much larger studies) will be needed. For this, collaboration between researchers as well as standardized protocols for phenotyping and DNA-collection will become increasingly important. Furthermore, understanding how the genes identified by GWAS contribute to ADHD risk, will require smart biological experiments facing the challenge of an organ as inaccessible and complex as the human brain.
References
Acosta MT, Arcos-Burgos M, Muenke M (2004) Attention deficit/hyperactivity disorder (ADHD): complex phenotype, simple genotype? Genet Med 6:1–15
Addamo PK, Farrow M, Hoy KE, Bradshaw JL, Georgiou-Karistianis N (2007) The effects of age and attention on motor overflow production—a review. Brain Res Rev 54:189–204
Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ (2004) Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem 88:1211–1219
Alcaraz WA, Gold DA, Raponi E, Gent PM, Concepcion D, Hamilton BA (2006) Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci USA 103:19424–19429
Allen-Brady K, Miller J, Matsunami N, Stevens J, Block H, Farley M, Krasny L, Pingree C, Lainhart J, Leppert M, McMahon WM, Coon H (2008) A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol Psychiatry. Feb 19 [Epub ahead of print]
Anisman H (2009) Cascading effects of stressors and inflammatory immune system activation: implications for major depressive disorder. J Psychiatry Neurosci 34:4–20
Anney RJ, Kenny E, O’Dushlaine CT, Lasky-Su J, Franke B, Morris DW, Neale BM, Asherson P, Faraone SV, Gill M (2008a) Non-random error in genotype calling procedures: implications for family-based and case–control genome-wide association studies. Am J Med Genet B Neuropsychiatr Genet 147B:1379–1386
Anney RJ, Lasky-Su J, O’Dushlaine C, Kenny E, Neale BM, Mulligan A, Franke B, Zhou K, Chen W, Christiansen H, Arias-Vasquez A, Banaschewski T, Buitelaar J, Ebstein R, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen H, Asherson P, Faraone SV, Gill M (2008b) Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet 147B:1369–1378
Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, Rapoport JL, Berg K, Bailey-Wilson JE, Muenke M (2004) Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet 75:998–1014
Arinami T, Ohtsuki T, Ishiguro H, Ujike H, Tanaka Y, Morita Y, Mineta M, Takeichi M, Yamada S, Imamura A, Ohara K, Shibuya H, Ohara K, Suzuki Y, Muratake T, Kaneko N, Someya T, Inada T, Yoshikawa T, Toyota T, Yamada K, Kojima T, Takahashi S, Osamu O, Shinkai T, Nakamura M, Fukuzako H, Hashiguchi T, Niwa SI, Ueno T, Tachikawa H, Hori T, Asada T, Nanko S, Kunugi H, Hashimoto R, Ozaki N, Iwata N, Harano M, Arai H, Ohnuma T, Kusumi I, Koyama T, Yoneda H, Fukumaki Y, Shibata H, Kaneko S, Higuchi H, Yasui-Furukori N, Numachi Y, Itokawa M, Okazaki Y (2005) Genomewide high-density SNP linkage analysis of 236 Japanese families supports the existence of schizophrenia susceptibility loci on chromosomes 1p, 14q, and 20p. Am J Hum Genet 77:937–944
Asherson P, Zhou K, Anney RJ, Franke B, Buitelaar J, Ebstein R, Gill M, Altink M, Arnold R, Boer F, Brookes K, Buschgens C, Butler L, Cambell D, Chen W, Christiansen H, Feldman L, Fleischman K, Fliers E, Howe-Forbes R, Goldfarb A, Heise A, Gabriels I, Johansson L, Lubetzki I, Marco R, Medad S, Minderaa R, Mulas F, Muller U, Mulligan A, Neale B, Rijsdijk F, Rabin K, Rommelse N, Sethna V, Sorohan J, Uebel H, Psychogiou L, Weeks A, Barrett R, Xu X, Banaschewski T, Sonuga-Barke E, Eisenberg J, Manor I, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Faraone SV (2008) A high-density SNP linkage scan with 142 combined subtype ADHD sib pairs identifies linkage regions on chromosomes 9 and 16. Mol Psychiatry 13(5):514–521
Bakker SC, van der Meulen EM, Buitelaar JK, Sandkuijl LA, Pauls DL, Monsuur AJ, van‘t Slot R, Minderaa RB, Gunning WB, Pearson PL, Sinke RJ (2003) A whole-genome scan in 164 Dutch sib pairs with attention-deficit/hyperactivity disorder: suggestive evidence for linkage on chromosomes 7p and 15q. Am J Hum Genet 72:1251–1260
Banerjee TD, Middleton F, Faraone SV (2007) Environmental risk factors for attention-deficit hyperactivity disorder. Acta Paediatr 96:1269–1274
Barkley RA, Smith KM, Fischer M, Navia B (2006) An examination of the behavioral and neuropsychological correlates of three ADHD candidate gene polymorphisms (DRD4 7 + , DBH TaqI A2, and DAT1 40 bp VNTR) in hyperactive and normal children followed to adulthood. Am J Med Genet B Neuropsychiatr Genet 141:487–498
Bazar KA, Yun AJ, Lee PY, Daniel SM, Doux JD (2006) Obesity and ADHD may represent different manifestations of a common environmental oversampling syndrome: a model for revealing mechanistic overlap among cognitive, metabolic, and inflammatory disorders. Med Hypotheses 66:263–269
Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V (2008) Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry 13:368–373
Biederman J, Kim JW, Doyle AE, Mick E, Fagerness J, Smoller JW, Faraone SV (2008) Sexually dimorphic effects of four genes (COMT, SLC6A2, MAOA, SLC6A4) in genetic associations of ADHD: a preliminary study. Am J Med Genet B Neuropsychiatr Genet 147B:1511–1518
Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Housman DE, Kazazian HH, Antonarakis S, Pulver AE (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73
Bobb AJ, Castellanos FX, Addington AM, Rapoport JL (2005) Molecular genetic studies of ADHD: 1991 to 2004. Am J Med Genet B Neuropsychiatr Genet 132:109–125
Brookes K, Xu X, Chen W, Zhou K, Neale B, Lowe N, Anney R, Franke B, Gill M, Ebstein R, Buitelaar J, Sham P, Campbell D, Knight J, Andreou P, Altink M, Arnold R, Boer F, Buschgens C, Butler L, Christiansen H, Feldman L, Fleischman K, Fliers E, Howe-Forbes R, Goldfarb A, Heise A, Gabriels I, Korn-Lubetzki I, Johansson L, Marco R, Medad S, Minderaa R, Mulas F, Muller U, Mulligan A, Rabin K, Rommelse N, Sethna V, Sorohan J, Uebel H, Psychogiou L, Weeks A, Barrett R, Craig I, Banaschewski T, Sonuga-Barke E, Eisenberg J, Kuntsi J, Manor I, McGuffin P, Miranda A, Oades RD, Plomin R, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Faraone SV, Asherson P (2006) The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry 11:934–953
Burton PR, Hansell AL, Fortier I, Manolio TA, Khoury MJ, Little J, Elliott P (2008) Size matters: just how big is BIG?: quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol 38(1):263–273
Cantwell DP (1975) Genetics of hyperactivity. J Child Psychol Psychiatry 16:261–264
Castellanos FX, Tannock R (2002) Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat Rev Neurosci 3:617–628
Chen W, Zhou K, Sham P, Franke B, Kuntsi J, Campbell D, Fleischman K, Knight J, Andreou P, Arnold R, Altink M, Boer F, Boholst MJ, Buschgens C, Butler L, Christiansen H, Fliers E, Howe-Forbes R, Gabriels I, Heise A, Korn-Lubetzki I, Marco R, Medad S, Minderaa R, Muller UC, Mulligan A, Psychogiou L, Rommelse N, Sethna V, Uebel H, McGuffin P, Plomin R, Banaschewski T, Buitelaar J, Ebstein R, Eisenberg J, Gill M, Manor I, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Taylor E, Thompson M, Faraone SV, Asherson P (2008) DSM-IV combined type ADHD shows familial association with sibling trait scores: a sampling strategy for QTL linkage. Am J Med Genet B Neuropsychiatr Genet 147B:1450–1460
Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, Badner JA, Matsui S, Conroy J, McQuaid D, Gergel J, Hatchwell E, Gilliam TC, Gershon ES, Nowak NJ, Dobyns WB, Cook EH Jr (2008) Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry 63:1111–1117
Collin RW, Kalay E, Tariq M, Peters T, van der Zwaag B, Venselaar H, Oostrik J, Lee K, Ahmed ZM, Caylan R, Li Y, Spierenburg HA, Eyupoglu E, Heister A, Riazuddin S, Bahat E, Ansar M, Arslan S, Wollnik B, Brunner HG, Cremers CW, Karaguzel A, Ahmad W, Cremers FP, Vriend G, Friedman TB, Riazuddin S, Leal SM, Kremer H (2008) Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet 82:125–138
Conners (2003) Conners’ rating scales—revised, 6th edn. MHS, Toronto
Craddock N, O’Donovan MC, Owen MJ (2008) Genome-wide association studies in psychiatry: lessons from early studies of non-psychiatric and psychiatric phenotypes. Mol Psychiatry 13:649–653
Craig IW, Harper E, Loat CS (2004) The genetic basis for sex differences in human behaviour: role of the sex chromosomes. Ann Hum Genet 68:269–284
de Silva MG, Elliott K, Dahl HH, Fitzpatrick E, Wilcox S, Delatycki M, Williamson R, Efron D, Lynch M, Forrest S (2003) Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J Med Genet 40:733–740
Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI Jr, Gershon ES (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 96:5604–5609
Deutsch CK, Matthysse S, Swanson JM, Farkas LG (1990) Genetic latent structure analysis of dysmorphology in attention deficit disorder. J Am Acad Child Adolesc Psychiatry 29:189–194
Devlin B, Bacanu SA, Klump KL, Bulik CM, Fichter MM, Halmi KA, Kaplan AS, Strober M, Treasure J, Woodside DB, Berrettini WH, Kaye WH (2002) Linkage analysis of anorexia nervosa incorporating behavioral covariates. Hum Mol Genet 11:689–696
Diamond A (2007) Consequences of variations in genes that affect dopamine in prefrontal cortex. Cereb Cortex 17(Suppl 1):i161–i170
Doyle AE, Willcutt EG, Seidman LJ, Biederman J, Chouinard VA, Silva J, Faraone SV (2005) Attention-deficit/hyperactivity disorder endophenotypes. Biol Psychiatry 57:1324–1335
Doyle AE, Ferreira MA, Sklar PB, Lasky-Su J, Petty C, Fusillo SJ, Seidman LJ, Willcutt EG, Smoller JW, Purcell S, Biederman J, Faraone SV (2008) Multivariate genomewide linkage scan of neurocognitive traits and ADHD symptoms: suggestive linkage to 3q13. Am J Med Genet B Neuropsychiatr Genet 147B:1399–1411
Dreher JC, Kohn P, Kolachana B, Weinberger DR, Berman KF (2008) Variation in dopamine genes influences responsivity of the human reward system. Proc Natl Acad Sci USA 106(2):617–622
Dudbridge F, Gusnanto A (2008) Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 32:227–234
Duggirala R, Almasy L, Blangero J (1999) Smoking behavior is under the influence of a major quantitative trait locus on human chromosome 5q. Genet Epidemiol 17(Suppl 1):S139–S144
Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH (2007) A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am J Psychiatry 164:656–662
Elia J, Devoto M (2007) ADHD genetics: 2007 update. Curr Psychiatry Rep 9:434–439
Faraone SV (2000) Genetics of childhood disorders: XX. ADHD, Part 4: is ADHD genetically heterogeneous? J Am Acad Child Adolesc Psychiatry 39:1455–1457
Faraone SV, Doyle AE (2001) The nature and heritability of attention-deficit/hyperactivity disorder. Child Adolesc Psychiatr Clin N Am 10:299–316
Faraone SV, Biederman J, Friedman D (2000a) Validity of DSM-IV subtypes of attention-deficit/hyperactivity disorder: a family study perspective. J Am Acad Child Adolesc Psychiatry 39:300–307
Faraone SV, Biederman J, Spencer T, Wilens T, Seidman LJ, Mick E, Doyle AE (2000b) Attention-deficit/hyperactivity disorder in adults: an overview. Biol Psychiatry 48:9–20
Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P (2005) Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry 57:1313–1323
Faraone SV, Doyle AE, Lasky-Su J, Sklar PB, D’Angelo E, Gonzalez-Heydrich J, Kratochvil C, Mick E, Klein K, Rezac AJ, Biederman J (2008) Linkage analysis of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 147B:1387–1391
Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, DeFelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, Maclean AW, St CD, Robinson M, Van BM, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Nicol FI, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N (2008) Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 40(9):1056–1058
Fliers E, Vermeulen S, Rijsdijk F, Altink M, Buschgens C, Rommelse N, Faraone S, Sergeant J, Buitelaar J, Franke B (2009) ADHD and poor motor performance from a family genetic perspective. J Am Acad Child Adolesc Psychiatry 48:25–34
Franke B, Hoogman M, Arias-Vasquez A, Heister JG, Savelkoul PJ, Naber M, Scheffer H, Kiemeney LA, Kan CC, Kooij JJ, Buitelaar JK (2008) Association of the dopamine transporter (SLC6A3/DAT1) gene 9-6 haplotype with adult ADHD. Am J Med Genet B Neuropsychiatr Genet 147B:1576–1579
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, DeFelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861
Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, Davidson S, Miller S, Fairburn C, Goodwin G, Neale MC, Fiddy S, Mott R, Allison DB, Flint J (2003) Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet 72:879–890
Galimberti D, Scarpini E, Venturelli E, Strobel A, Herterich S, Fenoglio C, Guidi I, Scalabrini D, Cortini F, Bresolin N, Lesch KP, Reif A (2008) Association of a NOS1 promoter repeat with Alzheimer’s disease. Neurobiol Aging 29:1359–1365
Garcia-Bueno B, Caso JR, Leza JC (2008) Stress as a neuroinflammatory condition in brain: damaging and protective mechanisms. Neurosci Biobehav Rev 32:1136–1151
Gottesman II, Gould TD (2003) The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160:636–645
Guimaraes AP, Zeni C, Polanczyk GV, Genro JP, Roman T, Rohde LA, Hutz MH (2007) Serotonin genes and attention deficit/hyperactivity disorder in a Brazilian sample: preferential transmission of the HTR2A 452His allele to affected boys. Am J Med Genet B Neuropsychiatr Genet 144B:69–73
Haberstick BC, Timberlake D, Hopfer CJ, Lessem JM, Ehringer MA, Hewitt JK (2008) Genetic and environmental contributions to retrospectively reported DSM-IV childhood attention deficit hyperactivity disorder. Psychol Med 38:1057–1066
Haefner S, Baghai TC, Schule C, Eser D, Spraul M, Zill P, Rupprecht R, Bondy B (2008) Impact of gene–gender effects of adrenergic polymorphisms on hypothalamic–pituitary–adrenal axis activity in depressed patients. Neuropsychobiology 58:154–162
Hebebrand J, Dempfle A, Saar K, Thiele H, Herpertz-Dahlmann B, Linder M, Kiefl H, Remschmidt H, Hemminger U, Warnke A, Knolker U, Heiser P, Friedel S, Hinney A, Schafer H, Nurnberg P, Konrad K (2006) A genome-wide scan for attention-deficit/hyperactivity disorder in 155 German sib-pairs. Mol Psychiatry 11:196–205
Heiser P, Heinzel-Gutenbrunner M, Frey J, Smidt J, Grabarkiewicz J, Friedel S, Kuhnau W, Schmidtke J, Remschmidt H, Hebebrand J (2006) Twin study on heritability of activity, attention, and impulsivity as assessed by objective measures. J Atten Disord 9:575–581
Holmans P, Zubenko GS, Crowe RR, DePaulo JR Jr, Scheftner WA, Weissman MM, Zubenko WN, Boutelle S, Murphy-Eberenz K, MacKinnon D, McInnis MG, Marta DH, Adams P, Knowles JA, Gladis M, Thomas J, Chellis J, Miller E, Levinson DF (2004) Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome 15q. Am J Hum Genet 74:1154–1167
Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, DePaulo JR Jr, Knowles JA, Zubenko WN, Murphy-Eberenz K, Marta DH, Boutelle S, McInnis MG, Adams P, Gladis M, Steele J, Miller EB, Potash JB, MacKinnon DF, Levinson DF (2007) Genetics of recurrent early-onset major depression (GenRED): final genome scan report. Am J Psychiatry 164:248–258
Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, Simonsen KL, Kirchgessner TG, Cannon CP, Devlin JJ, Braunwald E (2008a) Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. J Am Coll Cardiol 51:449–455
Iakoubova OA, Tong CH, Rowland CM, Kirchgessner TG, Young BA, Arellano AR, Shiffman D, Sabatine MS, Campos H, Packard CJ, Pfeffer MA, White TJ, Braunwald E, Shepherd J, Devlin JJ, Sacks FM (2008b) Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. J Am Coll Cardiol 51:435–443
Irmansyah XSSG, Handoko HY, Kusumawardhani A, Widyawati I, Amir N, Nasrun MW, Holmans P, Knapp M, Wildenauer DB (2008) Genome-wide scan in 124 Indonesian sib-pair families with schizophrenia reveals genome-wide significant linkage to a locus on chromosome 3p26-21. Am J Med Genet B Neuropsychiatr Genet 147B:1245–1252
Jacob CP, Romanos J, Dempfle A, Heine M, Windemuth-Kieselbach C, Kruse A, Reif A, Walitza S, Romanos M, Strobel A, Brocke B, Schafer H, Schmidtke A, Boning J, Lesch KP (2007) Co-morbidity of adult attention-deficit/hyperactivity disorder with focus on personality traits and related disorders in a tertiary referral center. Eur Arch Psychiatry Clin Neurosci 257:309–317
Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H (2001) A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA 98:585–590
Kendler KS (1998) Gender differences in the genetic epidemiology of major depression. J Gend Specif Med 1:28–31
Kessler RC, Adler L, Barkley R, Biederman J, Conners CK, Demler O, Faraone SV, Greenhill LL, Howes MJ, Secnik K, Spencer T, Ustun TB, Walters EE, Zaslavsky AM (2006) The prevalence and correlates of adult ADHD in the United States: results from the National Comorbidity Survey Replication. Am J Psychiatry 163:716–723
Khan SA, Faraone SV (2006) The genetics of ADHD: a literature review of 2005. Curr Psychiatry Rep 8:393–397
Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453:56–64
Kooij JJ, Buitelaar JK, van den Oord EJ, Furer JW, Rijnders CA, Hodiamont PP (2005) Internal and external validity of attention-deficit hyperactivity disorder in a population-based sample of adults. Psychol Med 35:817–827
Kuntsi J, Neale BM, Chen W, Faraone SV, Asherson P (2006) The IMAGE project: methodological issues for the molecular genetic analysis of ADHD. Behav Brain Funct 2:27
Lachman HM, Pedrosa E, Petruolo OA, Cockerham M, Papolos A, Novak T, Papolos DF, Stopkova P (2007) Increase in GSK3beta gene copy number variation in bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 144:259–265
Lang UE, Hellweg R, Bajbouj M, Gaus V, Sander T, Gallinat J (2008) Gender-dependent association of a functional NGF polymorphism with anxiety-related personality traits. Pharmacopsychiatry 41:196–199
Lange C, Blacker D, Laird NM (2004a) Family-based association tests for survival and times-to-onset analysis. Stat Med 23:179–189
Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM (2004b) PBAT: tools for family-based association studies. Am J Hum Genet 74:367–369
Lange C, van SK, Andrew T, Lyon H, DeMeo DL, Raby B, Murphy A, Silverman EK, MacGregor A, Weiss ST, Laird NM (2004c) A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat Appl Genet Mol Biol 3, Article 17
Lasky-Su J, Biederman J, Laird N, Tsuang M, Doyle AE, Smoller JW, Lange C, Faraone SV (2007) Evidence for an association of the dopamine D5 receptor gene on age at onset of attention deficit hyperactivity disorder. Ann Hum Genet 71:648–659
Lasky-Su J, Anney RJ, Neale BM, Franke B, Zhou K, Maller JB, Arias-Vasquez A, Chen W, Asherson P, Buitelaar J, Banaschewski T, Ebstein R, Gill M, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Taylor E, Daly M, Laird N, Lange C, Faraone SV (2008a) Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 147B:1355–1358
Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, Arias-Vasquez A, Chen W, Asherson P, Buitelaar J, Banaschewski T, Ebstein R, Gill M, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Taylor E, Daly M, Laird N, Lange C, Faraone SV (2008b) Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet 147B:1345–1354
Lerer B, Segman RH, Hamdan A, Kanyas K, Karni O, Kohn Y, Korner M, Lanktree M, Kaadan M, Turetsky N, Yakir A, Kerem B, Macciardi F (2003) Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol Psychiatry 8:488–498
Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schafer H, Walitza S, Reif A, Stephan DA, Jacob C (2008) Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm 115:1573–1585
Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O’Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O’Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet 73:34–48
Li D, Sham PC, Owen MJ, He L (2006) Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet 15:2276–2284
Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC (2008) Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry 13:407–416
Lu AT, Ogdie MN, Jarvelin MR, Moilanen IK, Loo SK, McCracken JT, McGough JJ, Yang MH, Peltonen L, Nelson SF, Cantor RM, Smalley SL (2008) Association of the cannabinoid receptor gene (CNR1) with ADHD and post-traumatic stress disorder. Am J Med Genet B Neuropsychiatr Genet 147B:1488–1494
Maher BS, Marazita ML, Moss HB, Vanyukov MM (1999) Segregation analysis of attention deficit hyperactivity disorder. Am J Med Genet 88:71–78
Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, Donnelly P, Faraone SV, Frazer K, Gabriel S, Gejman P, Guttmacher A, Harris EL, Insel T, Kelsoe JR, Lander E, McCowin N, Mailman MD, Nabel E, Ostell J, Pugh E, Sherry S, Sullivan PF, Thompson JF, Warram J, Wholley D, Milos PM, Collins FS (2007) New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet 39:1045–1051
Manolio TA, Brooks LD, Collins FS (2008) A HapMap harvest of insights into the genetics of common disease. J Clin Invest 118:1590–1605
Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW (2008) Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82:477–488
Maziade M, Roy MA, Chagnon YC, Cliche D, Fournier JP, Montgrain N, Dion C, Lavallee JC, Garneau Y, Gingras N, Nicole L, Pires A, Ponton AM, Potvin A, Wallot H, Merette C (2005) Shared and specific susceptibility loci for schizophrenia and bipolar disorder: a dense genome scan in Eastern Quebec families. Mol Psychiatry 10:486–499
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9:356–369
McLoughlin G, Ronald A, Kuntsi J, Asherson P, Plomin R (2007) Genetic support for the dual nature of attention deficit hyperactivity disorder: substantial genetic overlap between the inattentive and hyperactive–impulsive components. J Abnorm Child Psychol 35:999–1008
Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, Lipska BK (2008) Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res 98:129–138
Morrison JR, Stewart MA (1973) The psychiatric status of the legal families of adopted hyperactive children. Arch Gen Psychiatry 28:888–891
Morrison JR, Stewart MA (1974) Bilateral inheritance as evidence for polygenicity in the hyperactive child syndrome. J Nerv Ment Dis 158:226–228
Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, Al-Saad S, Ware J, Joseph RM, Greenblatt R, Gleason D, Ertelt JA, Apse KA, Bodell A, Partlow JN, Barry B, Yao H, Markianos K, Ferland RJ, Greenberg ME, Walsh CA (2008) Identifying autism loci and genes by tracing recent shared ancestry. Science 321:218–223
Neale BM, Purcell S (2008) The positives, protocols, and perils of genome-wide association. Am J Med Genet B Neuropsychiatr Genet 147B:1288–1294
Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, Arias-Vasquez A, Asherson P, Chen W, Banaschewski T, Buitelaar J, Ebstein R, Gill M, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Sonuga-Barke E, Mulas F, Taylor E, Laird N, Lange C, Daly M, Faraone SV (2008a) Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 147B:1337–1344
Neale BM, Sham PC, Purcell S, Banaschewski T, Buitelaar J, Franke B, Sonuga-Barke E, Ebstein R, Eisenberg J, Mulligan A, Gill M, Manor I, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Chen W, Zhou K, Asherson P, Faraone SV (2008b) Population differences in the International Multi-Centre ADHD Gene Project. Genet Epidemiol 32:98–107
O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Gejman PV, Sanders AR, Duan J, Levinson DF, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ (2008) Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 40(9):1053–1055
O’Donovan MC, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Giegling I, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Adolfsson R, Osby U, Terenius L, Jonsson EG, Cichon S, Nothen MM, Gill M, Corvin AP, Rujescu D, Gejman PV, Kirov G, Craddock N, Williams NM, Owen MJ (2009) Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Mol Psychiatry 14:30–36
Ogdie MN, Macphie IL, Minassian SL, Yang M, Fisher SE, Francks C, Cantor RM, McCracken JT, McGough JJ, Nelson SF, Monaco AP, Smalley SL (2003) A genomewide scan for attention-deficit/hyperactivity disorder in an extended sample: suggestive linkage on 17p11. Am J Hum Genet 72:1268–1279
Ogdie MN, Fisher SE, Yang M, Ishii J, Francks C, Loo SK, Cantor RM, McCracken JT, McGough JJ, Smalley SL, Nelson SF (2004) Attention deficit hyperactivity disorder: fine mapping supports linkage to 5p13, 6q12, 16p13, and 17p11. Am J Hum Genet 75:661–668
Ogdie MN, Bakker SC, Fisher SE, Francks C, Yang MH, Cantor RM, Loo SK, van der Meulen E, Pearson P, Buitelaar J, Monaco A, Nelson SF, Sinke RJ, Smalley SL (2006) Pooled genome-wide linkage data on 424 ADHD ASPs suggests genetic heterogeneity and a common risk locus at 5p13. Mol Psychiatry 11:5–8
Oh I, Shimizu H, Satoh T, Okada S, Adachi S, Inoue K, Eguchi H, Yamamoto M, Imaki T, Hashimoto K, Tsuchiya T, Monden T, Horiguchi K, Yamada M, Mori M (2006) Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature 443:709–712
Okamura N, Hashimoto K, Iyo M, Shimizu E, Dempfle A, Friedel S, Reinscheid RK (2007) Gender-specific association of a functional coding polymorphism in the Neuropeptide S receptor gene with panic disorder but not with schizophrenia or attention-deficit/hyperactivity disorder. Prog Neuropsychopharmacol Biol Psychiatry 31:1444–1448
Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ (2006) Common Kibra alleles are associated with human memory performance. Science 314:475–478
Patel SD, Chen CP, Bahna F, Honig B, Shapiro L (2003) Cadherin-mediated cell–cell adhesion: sticking together as a family. Curr Opin Struct Biol 13:690–698
Pe’er I, Yelensky R, Altshuler D, Daly MJ (2008) Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 32:381–385
Pearson TA, Manolio TA (2008) How to interpret a genome-wide association study. JAMA 299:1335–1344
Penzes P, Jones KA (2008) Dendritic spine dynamics—a key role for kalirin-7. Trends Neurosci 31:419–427
Polanczyk G, Rohde LA (2007) Epidemiology of attention-deficit/hyperactivity disorder across the lifespan. Curr Opin Psychiatry 20:386–392
Ponce G, Hoenicka J, Rubio G, Ampuero I, Jimenez-Arriero MA, Rodriguez-Jimenez R, Palomo T, Ramos JA (2003) Association between cannabinoid receptor gene (CNR1) and childhood attention deficit/hyperactivity disorder in Spanish male alcoholic patients. Mol Psychiatry 8:466–467
Psychiatric GWAS Consortium Steering Committee (2009) A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry 14:10–17
Rasmussen ER, Neuman RJ, Heath AC, Levy F, Hay DA, Todd RD (2004) Familial clustering of latent class and DSM-IV defined attention-deficit/hyperactivity disorder (ADHD) subtypes. J Child Psychol Psychiatry 45:589–598
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME (2006) Global variation in copy number in the human genome. Nature 444:444–454
Reif A, Herterich S, Strobel A, Ehlis AC, Saur D, Jacob CP, Wienker T, Topner T, Fritzen S, Walter U, Schmitt A, Fallgatter AJ, Lesch KP (2006) A neuronal nitric oxide synthase (NOS-I) haplotype associated with schizophrenia modifies prefrontal cortex function. Mol Psychiatry 11:286–300
Reif A, Jacob CP, Rujescu D, Herterich S, Lang S, Gutknecht L, Baehne CG, Strobel A, Freitag CM, Giegling I, Romanos M, Hartmann A, Rosler M, Renner TJ, Fallgatter AJ, Retz W, Ehlis AC, Lesch KP (2009) Influence of functional variant of neuronal nitric oxide synthase on impulsive behaviors in humans. Arch Gen Psychiatry 66:41–50
Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517
Romanos M, Freitag C, Jacob C, Craig DW, Dempfle A, Nguyen TT, Halperin R, Walitza S, Renner TJ, Seitz C, Romanos J, Palmason H, Reif A, Heine M, Windemuth-Kieselbach C, Vogler C, Sigmund J, Warnke A, Schafer H, Meyer J, Stephan DA, Lesch KP (2008) Genome-wide linkage analysis of ADHD using high-density SNP arrays: novel loci at 5q13.1 and 14q12. Mol Psychiatry 13:522–530
Rommelse NN (2008) Endophenotypes in the genetic research of ADHD over the last decade: have they lived up to their expectations? Expert Rev Neurother 8:1425–1429
Rommelse NN, Altink ME, Arias-Vasquez A, Buschgens CJ, Fliers E, Faraone SV, Buitelaar JK, Sergeant JA, Franke B, Oosterlaan J (2008a) A review and analysis of the relationship between neuropsychological measures and DAT1 in ADHD. Am J Med Genet B Neuropsychiatr Genet 147B:1536–1546
Rommelse NN, Altink ME, Arias-Vasquez A, Buschgens CJ, Fliers E, Faraone SV, Buitelaar JK, Sergeant JA, Oosterlaan J, Franke B (2008b) Differential association between MAOA, ADHD and neuropsychological functioning in boys and girls. Am J Med Genet B Neuropsychiatr Genet 147B:1524–1530
Rommelse NN, Arias-Vasquez A, Altink ME, Buschgens CJ, Fliers E, Asherson P, Faraone SV, Buitelaar JK, Sergeant JA, Oosterlaan J, Franke B (2008c) Neuropsychological endophenotype approach to genome-wide linkage analysis identifies susceptibility loci for ADHD on 2q21.1 and 13q12.11. Am J Hum Genet 83:99–105
Rosler M, Retz W, Retz-Junginger P, Thome J, Supprian T, Nissen T, Stieglitz RD, Blocher D, Hengesch G, Trott GE (2004) Tools for the diagnosis of attention-deficit/hyperactivity disorder in adults. Self-rating behaviour questionnaire and diagnostic checklist. Nervenarzt 75:888–895
Rucci P, Nimgaonkar VL, Mansour H, Miniati M, Masala I, Fagiolini A, Cassano GB, Frank E (2009) Gender moderates the relationship between mania spectrum and serotonin transporter polymorphisms in depression. Am J Med Genet B Neuropsychiatr Genet. Jan 5 [Epub ahead of print]
Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, Agrawal A, Dick DM, Heath AC, Todorov AA, Maunu H, Heikkila K, Morley KI, Rice JP, Todd RD, Kaprio J, Peltonen L, Martin NG, Goate AM, Madden PA (2007) Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet 80:856–866
Schultz MR, Rabi K, Faraone SV, Kremen W, Lyons MJ (2006) Efficacy of retrospective recall of attention-deficit hyperactivity disorder symptoms: a twin study. Twin Res Hum Genet 9:220–232
Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M (2004) Large-scale copy number polymorphism in the human genome. Science 305:525–528
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M (2007) Strong association of de novo copy number mutations with autism. Science 316:445–449
Serretti A, Olgiati P, De RD (2007) Genetics of Alzheimer’s disease. A rapidly evolving field. J Alzheimers Dis 12:73–92
Shiffman D, Chasman DI, Zee RY, Iakoubova OA, Louie JZ, Devlin JJ, Ridker PM (2008) A kinesin family member 6 variant is associated with coronary heart disease in the Women’s Health Study. J Am Coll Cardiol 51:444–448
Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, DeFelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, MacIntyre DJ, McLean A, VanBeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM (2008) Whole-genome association study of bipolar disorder. Mol Psychiatry 13:558–569
Smalley SL, McGough JJ, Del’Homme M, NewDelman J, Gordon E, Kim T, Liu A, McCracken JT (2000) Familial clustering of symptoms and disruptive behaviors in multiplex families with attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 39:1135–1143
Sonuga-Barke EJ, Lasky-Su J, Neale BM, Oades R, Chen W, Franke B, Buitelaar J, Banaschewski T, Ebstein R, Gill M, Anney R, Miranda A, Mulas F, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Thompson M, Asherson P, Faraone SV (2008) Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet 147B:1359–1368
Spielman RS, Ewens WJ (1996) The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet 59:983–989
Sprich S, Biederman J, Crawford MH, Mundy E, Faraone SV (2000) Adoptive and biological families of children and adolescents with ADHD. J Am Acad Child Adolesc Psychiatry 39:1432–1437
Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, Gunnarsdottir S, Ivarsson O, Chou TT, Hjaltason O, Birgisdottir B, Jonsson H, Gudnadottir VG, Gudmundsdottir E, Bjornsson A, Ingvarsson B, Ingason A, Sigfusson S, Hardardottir H, Harvey RP, Lai D, Zhou M, Brunner D, Mutel V, Gonzalo A, Lemke G, Sainz J, Johannesson G, Andresson T, Gudbjartsson D, Manolescu A, Frigge ML, Gurney ME, Kong A, Gulcher JR, Petursson H, Stefansson K (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892
Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di FM, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St CD, Stefansson K (2008) Large recurrent microdeletions associated with schizophrenia. Nature 455:232–236
Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL (2008) Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry 13:570–584
Szatmari P, Maziade M, Zwaigenbaum L, Merette C, Roy MA, Joober R, Palmour R (2007a) Informative phenotypes for genetic studies of psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 144B:581–588
Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijimans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le CA, Berney T, McConachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A (2007b) Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39:319–328
Szyf M, Weaver I, Meaney M (2007) Maternal care, the epigenome and phenotypic differences in behavior. Reprod Toxicol 24:9–19
Szyf M, McGowan P, Meaney MJ (2008) The social environment and the epigenome. Environ Mol Mutagen 49:46–60
Takeuchi T, Ohtsuki Y (2001) Recent progress in T-cadherin (CDH13, H-cadherin) research. Histol Histopathol 16:1287–1293
Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, Ohtsuki Y (2000) Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem 74:1489–1497
Taylor E, Schachar R, Thorley G, Wieselberg M (1986) Conduct disorder and hyperactivity: I. Separation of hyperactivity and antisocial conduct in British child psychiatric patients. Br J Psychiatry 149:760–767
Thapar A, Langley K, O’Donovan M, Owen M (2006) Refining the attention deficit hyperactivity disorder phenotype for molecular genetic studies. Mol Psychiatry 11:714–720
The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320
Tom TY, Emtage P, Funk WD, Hu T, Arterburn M, Park EE, Rupp F (2004) TAFA: a novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics 83:727–734
Trikalinos TA, Karvouni A, Zintzaras E, Ylisaukko-oja T, Peltonen L, Jarvela I, Ioannidis JP (2006) A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry 11:29–36
Uhl GR, Drgon T, Johnson C, Li CY, Contoreggi C, Hess J, Naiman D, Liu QR (2008a) Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify “connectivity constellation” and drug target genes with pleiotropic effects. Ann N Y Acad Sci 1141:318–381
Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, Harano M, Sekine Y, Inada T, Ozaki N, Iyo M, Iwata N, Yamada M, Sora I, Chen CK, Liu HC, Ujike H, Lin SK (2008b) Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry 65:345–355
Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos AC, Banna L, Brereton AV, Hill A, Bisgaard AM, Muller I, Hultschig C, Erdogan F, Wieczorek G, Ropers HH (2007) Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat 28:674–682
Valera EM, Faraone SV, Murray KE, Seidman LJ (2007) Meta-analysis of structural imaging findings in attention-deficit/hyperactivity disorder. Biol Psychiatry 61:1361–1369
van Es MA, Van Vught PW, Blauw HM, Franke L, Saris CG, Andersen PM, Van Den Bosch L, de Jong SW, van’t Slot R, Birve A, Lemmens R, de Jong V, Baas F, Schelhaas HJ, Sleegers K, Van Broeckhoven C, Wokke JH, Wijmenga C, Robberecht W, Veldink JH, Ophoff RA, van den Berg LH (2007) ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol 6:869–877
van Haren NE, Bakker SC, Kahn RS (2008) Genes and structural brain imaging in schizophrenia. Curr Opin Psychiatry 21:161–167
van Steen K, Lange C (2005) PBAT: a comprehensive software package for genome-wide association analysis of complex family-based studies. Hum Genomics 2:67–69
Vansteelandt S, DeMeo DL, Lasky-Su J, Smoller JW, Murphy AJ, McQueen M, Schneiter K, Celedon JC, Weiss ST, Silverman EK, Lange C (2008) Testing and estimating gene–environment interactions in family-based association studies. Biometrics 64:458–467
Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE (2007) High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet 81:1232–1250
Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE (2008) A functional genetic link between distinct developmental language disorders. N Engl J Med 359:2337–2345
Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, van Geurts KA, Brunner HG, Ophoff RA, Veltman JA (2008) Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet 83:504–510
Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J (2008) Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320:539–543
Walters JT, Owen MJ (2007) Endophenotypes in psychiatric genetics. Mol Psychiatry 12:886–890
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ (2008) Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358:667–675
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678
Willcutt EG, Pennington BF, DeFries JC (2000) Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. Am J Med Genet 96:293–301
Xiang M, Mohamalawari D, Rao R (2005) A novel isoform of the secretory pathway Ca2+, Mn(2+)-ATPase, hSPCA2, has unusual properties and is expressed in the brain. J Biol Chem 280:11608–11614
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341
Zhou K, Asherson P, Sham P, Franke B, Anney RJ, Buitelaar J, Ebstein R, Gill M, Brookes K, Buschgens C, Campbell D, Chen W, Christiansen H, Fliers E, Gabriels I, Johansson L, Marco R, Mulas F, Muller U, Mulligan A, Neale BM, Rijsdijk F, Rommelse N, Uebel H, Psychogiou L, Xu X, Banaschewski T, Sonuga-Barke E, Eisenberg J, Manor I, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Taylor E, Thompson M, Faraone SV (2008a) Linkage to chromosome 1p36 for attention-deficit/hyperactivity disorder traits in school and home settings. Biol Psychiatry 64:571–576
Zhou K, Chen W, Buitelaar J, Banaschewski T, Oades RD, Franke B, Sonuga-Barke E, Ebstein R, Eisenberg J, Gill M, Manor I, Miranda A, Mulas F, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Lasky-Su J, Taylor E, Brookes KJ, Xu X, Neale BM, Rijsdijk F, Thompson M, Asherson P, Faraone SV (2008b) Genetic heterogeneity in ADHD: DAT1 gene only affects probands without CD. Am J Med Genet B Neuropsychiatr Genet 147B:1481–1487
Zhou K, Dempfle A, Arcos-Burgos M, Bakker SC, Banaschewski T, Biederman J, Buitelaar J, Castellanos FX, Doyle A, Ebstein RP, Ekholm J, Forabosco P, Franke B, Freitag C, Friedel S, Gill M, Hebebrand J, Hinney A, Jacob C, Lesch KP, Loo SK, Lopera F, McCracken JT, McGough JJ, Meyer J, Mick E, Miranda A, Muenke M, Mulas F, Nelson SF, Nguyen TT, Oades RD, Ogdie MN, Palacio JD, Pineda D, Reif A, Renner TJ, Roeyers H, Romanos M, Rothenberger A, Schafer H, Sergeant J, Sinke RJ, Smalley SL, Sonuga-Barke E, Steinhausen HC, van der Meulen E, Walitza S, Warnke A, Lewis CM, Faraone SV, Asherson P (2008c) Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 147B:1392–1398
Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J (2007) CNR1 variation modulates risk for drug and alcohol dependence. Biol Psychiatry 62:616–626
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Franke, B., Neale, B.M. & Faraone, S.V. Genome-wide association studies in ADHD. Hum Genet 126, 13–50 (2009). https://doi.org/10.1007/s00439-009-0663-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-009-0663-4