Abstract
A multiple sclerosis (MS) lesion at an early stage undergoes active blood brain barrier (BBB) breakdown. Identifying MS lesions in a patient which are undergoing active BBB breakdown is of critical importance for MS burden evaluation and treatment planning. However in non-contrast enhanced structural magnetic resonance imaging (MRI) the regions of the lesion undergoing active BBB breakdown cannot be distinguished from the other parts of the lesion. Hence gadolinium (Gd) contrast enhanced T1-weighted MR images are used for this task. However some side effects of Gd injection into patients have been increasingly reported recently. The BBB breakdown is reflected by the condition of tissue microstructure such as increased inflammation, presence of higher extra-cellular matter and debris. We thus propose a framework to predict enhancing regions in MS lesions using tissue microstructure information derived from T2 relaxometry and diffusion MRI (dMRI) multi-compartment models. We show that combination of the dMRI and T2 relaxometry microstructure information can distinguish the Gd enhancing lesion regions from the other regions in MS lesions.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Multiple sclerosis (MS) patients have multiple focal lesions in the brain. In the early stages, the MS lesions undergo active blood brain barrier (BBB) breakdown [1, 2]. Lesions in this stage are referred to as enhancing lesions and the clinical significance of its identification is well established. Gadolinium based contrast agents (GBCA) are popularly used by radiologists to identify enhancing MS lesions. A T1-weighted MRI acquired post GBCA injection is a part of the recommended MRI protocols for diagnosis and follow-up examinations of MS patients [3]. However, the use of GBCAs has been a recent topic of debate, primarily due to reports of Gd deposition in the brain [4, 5]. Suggestions insisting on greater debate before GBCA administration has gained traction due to observed MRI signal changes in the brain tissues due to repeated GBCA administrations and other possible health issues [6]. A MS lesions in its enhancing stage has different pathological traits as compared to late or non-enhancing MS lesions [2]. Regions of lesion undergoing active BBB breakdown has a higher water content owing to the undergoing tissue damages such as functional impairment of the morphologically intact endothelial cells [1]. Hence by the virtue of the brain tissue microstructure characteristics of MS lesions, we may identify enhancing regions in the lesion (if any). Although MRI effectively provides in-vivo images of the brain, it is constrained by the limited imaging resolution it can provide. This limitation is primarily attributed to hardware limits and the need for adhering to reasonable scan times for clinical implementations. However, advanced MRI methods, such as diffusion MRI (dMRI) and T2 relaxometry help us obtain estimates on condition of brain tissue microstructure. The multi-compartment models (MCM) [7] in dMRI provide information on the organization of the nerve fibers in the brain. MS lesions disrupt the normal organization of fibers in the brain. The extent of this damage may be assessed from the tissue microstrucutre estimates derived from dMRI MCMs. Myelin is also a critical biomarker in neurodegenerative diseases such as MS [2, 8, 9]. Demyelination marks the onset of MS [2]. Myelin has a very short T2 relaxation time (<50 ms) due to its tightly wrapped structure [10]. Due to higher TEs in dMRI, the myelin information is not present in the dMRI signals. However, myelin information can be obtained by estimating the myelin water fraction (MWF) from T2 relaxometry MRI signal [8, 9]. The inflammation in MS lesions can be assessed from T2 relaxometry and dMRI signal. Hence, by combining the tissue microstructure information from these two MRI methods, we can obtain considerable information on brain tissue health. In this work we combine the microstructure information obtained from T2 relaxometry and dMRI to identify Gd enhanced regions in MS lesions. We performed experiments to evaluate whether combining the tissue microstructures is advantageous as compared to using them alone. The observations from this experiment is carried forward to the next stage where we perform enhancing lesion region predictions in a MS patient.
2 Materials and Methods
2.1 Multi-Compartment T2 Relaxometry Model (MCT2)
Three T2 relaxometry compartments are considered in a voxel based on their T2 relaxations times and are referred to as short-, medium- and high-T2. The short-T2 compartment conveys information on brain tissues with T2 relaxation times shorter than 50 ms. These tissues primarily include myelin and highly myelinated axons [10]. The high-T2 compartment represents the tissues with T2 values greater than 1000ms, comprising primarily of free fluids (CSF in healthy volunteer) and water accumulated in tissues due to pathology (edema regions in MS lesions). The medium-T2 compartment is a mixed pool and conveys information on intracellular matter (such as unmyelinated axons and glia), intra and extracellular fluids [10]. The T2 space is modeled as a weighted mixture of the three compartments, where each compartment is represented by a continuous probability density function (PDF). The signal of a voxel at the i-th echo in the MCT2 is modeled as:
In Eq. (1), \(f_j \left( T_{2}; \mathbf {p} \right) \) is the chosen PDF to represent the j-th compartment with parameters \(\mathbf {p_j}=\lbrace p_{j_1},p_{j_2},\ldots \rbrace \). We used the 2D multislice Carr-Purcell-Meiboom-Gill (CPMG) sequence to acquire T2 relaxometry data. CPMG sequences suffer from the effect of the stimulated echoes due to imperfect refocusing. It is important to address this effect as this leads to errors in T2 estimation [11]. Here we tackle is the problem of stimulated echoes using the iterative technique of Extended Phase Graph (EPG) algorithm [12]. Each compartment’s weight is obtained as, \(w_j=\alpha _j / \sum _i \alpha _i\). Simultaneous estimation of the weights and parameters of the distributions of such multi-compartment models is non-trivial and not reliable in terms of robustness and accuracy [13]. Hence we choose to fix the PDF parameters. In this work, the \(\lbrace f_j(\cdot ) \rbrace _{j=1}^{3}\) are chosen as Gaussian PDFs. Their mean and standard deviation are fixed based on the findings from the literature [8,9,10] and are set as \(\mu =\lbrace 20,100,2000 \rbrace \) and \(\sigma = \lbrace 5,10,80 \rbrace \) (all values in milliseconds). Estimating the model thus resorts to finding the optimal \(\left\{ {\alpha _j}\right\} _{j=1}^{3}\) and \(B_1\) for the following least squares problem:
where \(\mathbf {Y}\in \mathbb {R}^m\) is the observed signal; m is the number of echoes; \(\mathbf {\alpha }\in \mathbb {R^+}^3\). Although the optimization of \(\mathbf {\alpha }\) and \(B_1\) are linear and non-linear in nature, these variables are linearly separable. \(\alpha \) and \(B_1\) are computed by non-negative least squares and BOBYQA optimization respectively. The weights of short-T2 (\(w_s\)), medium-T2 (\(w_m\)) and high-T2 (\(w_h\)) compartment for every voxel are used as a feature for each voxel.
2.2 Multi-Compartment Diffusion Model (MCDiff)
For diffusion MRI (dMRI), we considered the recently introduced MCM as they provide an intuitive way of describing the different fascicles, cells and free water contributions to each voxel. MCM are defined as a weighted sum of several compartments each describing a fascicle (i.e. a dense set of fibers sharing the same orientation) or isotropic matter (such as free water or water trapped in cell bodies). Similar to T2 relaxometry, each compartment in MCDiff is defined by a PDF. However in MCDiff, the PDFs describe water diffusion probability inside the compartments. A variety of compartment types may be defined [7] based on the assumed white matter microstructure and the acquired dMRI data (the more gradient directions and b-values, the more information may be extracted). Our proposed method is quite independent of this choice and can be applied generically to all parameters that may be extracted from MCM. In the specific study in Sect. 3, we have focused on the following model:
where \(f_w + \sum _i a_i = 1\) are the weights of the individual compartments, \(p_{FW}\) is an isotropic Gaussian PDF specific to free water (i.e. with a variance of \(3.0\times 10^{-3}\,\mathrm{mm}^{2}\,\mathrm{s}^{-1}\)), \(p_i(x)\) denotes the i-th fascicle compartment PDF here defined as a stick model (i.e. a Gaussian PDF with equal secondary eigenvalues, fixed from an outside reference). We have specifically chosen this ball and stick model for our experiments as it can be estimated reliably on our clinical data (see Sect. 2.3). This estimation was performed using the method proposed by Stamm et al. [14], which uses a variable projection on the linear elements of the cost function (the compartment weights) to perform a fast maximum likelihood estimation of the model parameters with Levenberg-Marquardt optimization. We then defined different parameters from this MCM which describe the white matter microstructure inside the voxel. First, each anisotropic compartment \(p_i\) is defined as a constrained tensor. Hence we can extract the usual tensor scalar maps for each anisotropic compartment. However, to enable comparison between voxels, we need to average those compartment specific values over all anisotropic compartments. We have thus computed the weighted average of those values (using the weights \(a_i\)) to get the following scalar maps: fractional anisotropy \(\left( FA_{mc}\right) \), apparent diffusion coefficient \(\left( ADC_{mc}\right) \) and axial diffusivity \(\left( AD_{mc}\right) \). In addition to those maps, the weight of isotropic free water \(\left( f_w\right) \) is a crucial one that could identify edema or other free water related phenomena and we therefore included it in the parameters as well.
2.3 Data
All acquisitions were made on a 3T MRI scanner. The T2 relaxometry data was obtained using a 2D multislice CPMG sequence with the following specifications: first echo time (TE) = 13.8 ms; echo spacing = 13.8 ms; 7 echoes; repetition time (TR) = 4530 ms; \(1.33\times 1.33\times 3\,\mathrm{mm}^{3}\) voxel resolution; acquisition time was just less than 7 min. The dMRI acquisition was performed with 30 directions on a single shell of b-value at \(1000\,\mathrm{s/mm}^{2}\), with a \(2\times 2\times 2\,\mathrm{mm}^{3}\) voxel resolution, on a \(128\times 128\times 60\) matrix with TE and TR of 94 ms and 9.3 s respectively. Transverse SE T1-w images (\(1\times 1\times 3\,\mathrm{mm}^{3}\)) post Gd contrast agent infusion (0.1 mmol/kg gadopentetate dimeglumine) were acquired to find Gd enhanced lesions. A T1-w image was also acquired for performing the distortion correction in diffusion images. A 3D MPRAGE image was acquired with inversion time, TR and TE of 900 ms, 1900 ms and 2.98 ms respectively. The voxel resolution was \(1 \times 1 \times 1\,\mathrm{mm}^{3}\) on a \(256 \times 256 \times 160\) matrix. Lesions were segmented on T2-w images by radiologist. Hence all images were registered to the T2 image (\(1\times 1\times 3\,\mathrm{mm}^{3}\) voxel resolution) linear registration using a block-matching algorithm [15, 16]. Our data set consisted of 10 MS patient datasets demonstrating clinically isolated syndrome (CIS) condition. There were a total of 227 MS lesions in all patients, out of which 28 lesions had gadolinium enhancing regions. The voxels are divided into two groups: (a) (E+): voxels appearing on Gd enhanced T1 SE images and (b) (L−): lesion voxels which are hyperintense on T2-w images but do not appear on Gd enhanced T1-weighted images. The protocols were approved by the institutional review board, and all participants gave their written consent.
2.4 Identifying Enhancing Voxels in Lesions
We performed enhancing voxel identification using the MCT2 and MCDiff estimates. In our database, we had 15012 and 3904 (L−) and (E+) voxels respectively. We adopted a random shuffle and repeat strategy to compensate for the imbalance in the class. 5000 (L−) and 3400 (E+) voxels are randomly selected from the dataset to train the classifier. The remaining (L−) and (E+) voxels are then used to evaluate the classifier performance. This method is repeated 100 times to avoid any bias in sampling the data set for model training. The accuracy statistics are recorded for every repetition. It shall be noted that the model from one repetition is not retained for the next. For a new repetition, the model is trained and validated on a different dataset. We then observe the validation error of the classifier over 100 repitions. We used support vector machine classifier (with radial basis function kernel) in this work [17].
 The predictions are performed using three features sets: (a) MCT2 derived microstructure information: \(\mathcal {F}_R= \left\{ w_s,\right. w_m, \left. w_h,\right\} \in \mathbb {R}^{3}\), (b) MDiff derived microstructure information: \(\mathcal {F}_D= \left\{ {f_w}\right. , FA_{mc}, ADC_{mc}, \left. AD_{mc}\right\} \in \mathbb {R}^{4}\) and (c) a features set containing both MCT2 and MCDiff derived microstructure information \(\left( \mathcal {F}_{RD}\in \mathbb {R}^{7} \right) \). The aim of this experiment is to observe whether combining the diffusion and T2 relaxometry derived microstructure increases the accuracy of prediction. The observations from this experiment will help us comment on complimentary nature of the feature sets (if any).
The predictions are performed using three features sets: (a) MCT2 derived microstructure information: \(\mathcal {F}_R= \left\{ w_s,\right. w_m, \left. w_h,\right\} \in \mathbb {R}^{3}\), (b) MDiff derived microstructure information: \(\mathcal {F}_D= \left\{ {f_w}\right. , FA_{mc}, ADC_{mc}, \left. AD_{mc}\right\} \in \mathbb {R}^{4}\) and (c) a features set containing both MCT2 and MCDiff derived microstructure information \(\left( \mathcal {F}_{RD}\in \mathbb {R}^{7} \right) \). The aim of this experiment is to observe whether combining the diffusion and T2 relaxometry derived microstructure increases the accuracy of prediction. The observations from this experiment will help us comment on complimentary nature of the feature sets (if any).
 In this experiment we illustrate the application of the proposed method on a MS patient. We maintain a MS patient dataset which was never used for training the data set in any of the repetitions. The classifier trained in every repetition is used to predict to which category the voxels in the validation image belong. We perform a majority voting on the 100 predictions to decide the final prediction for each voxel. Subsequently we compute the dice measure on the (E+) and (L−) masks to judge the performance of the classifier. The classifier implementation was performed using the scikit-learn package in Python v2.7.
In this experiment we illustrate the application of the proposed method on a MS patient. We maintain a MS patient dataset which was never used for training the data set in any of the repetitions. The classifier trained in every repetition is used to predict to which category the voxels in the validation image belong. We perform a majority voting on the 100 predictions to decide the final prediction for each voxel. Subsequently we compute the dice measure on the (E+) and (L−) masks to judge the performance of the classifier. The classifier implementation was performed using the scikit-learn package in Python v2.7.
3 Results
 We show in Fig. 1 comparison of the prediction performance of the classifier on validation sets over 100 repititions using features sets derived from: (a) MCT2 model only \(\left( \mathcal {F}_R\right) \), (b) MCDiff model only \(\left( \mathcal {F}_D\right) \) and (c) combination of both \(\left( \mathcal {F}_{RD}\right) \). The mean overall accuracy of prediction when using \(\mathcal {F}_{RD}\), \(\mathcal {F}_R\) and \(\mathcal {F}_D\) are 85.57%, 84.24% and 81.73% respectively. From the overall accuracy plot shown in Fig. 1 we observe that combining the microstructure measures from MCT2 and MCDiff model yields better (E+) detection. The true positive rate (TPR) and true negative rate (TNR) plots in Fig. 1 show that MCT2 and MCDiff features are better than the other at detecting non-enhanced and enhanced voxels respectively in MS lesions. However, combining both features \(\left( \mathcal {F}_{RD}\right) \) yields better prediction results. Hence we use \(\mathcal {F}_{RD}\) for performing predictions in experiment-2.
We show in Fig. 1 comparison of the prediction performance of the classifier on validation sets over 100 repititions using features sets derived from: (a) MCT2 model only \(\left( \mathcal {F}_R\right) \), (b) MCDiff model only \(\left( \mathcal {F}_D\right) \) and (c) combination of both \(\left( \mathcal {F}_{RD}\right) \). The mean overall accuracy of prediction when using \(\mathcal {F}_{RD}\), \(\mathcal {F}_R\) and \(\mathcal {F}_D\) are 85.57%, 84.24% and 81.73% respectively. From the overall accuracy plot shown in Fig. 1 we observe that combining the microstructure measures from MCT2 and MCDiff model yields better (E+) detection. The true positive rate (TPR) and true negative rate (TNR) plots in Fig. 1 show that MCT2 and MCDiff features are better than the other at detecting non-enhanced and enhanced voxels respectively in MS lesions. However, combining both features \(\left( \mathcal {F}_{RD}\right) \) yields better prediction results. Hence we use \(\mathcal {F}_{RD}\) for performing predictions in experiment-2.
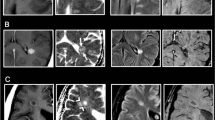
 Results are shown in Fig. 2. This MS patient had 18 MS lesions out of which 3 of them had Gd enhancing regions. The dice score for (E+) and (L−) voxel prediction was 0.64 and 0.86 respectively. The top row in Fig. 2 shows a lesion which had only (L−) voxels and the proposed method successfully predicted that there were no (E+) voxels in the lesion. The second and third row of Fig. 2 shows performance of the method in presence of Gd enhanced voxels in the lesion. Our method identified the (E+) voxels in the lesion. However there were false positives around the Gd enhanced core of the lesions where non-enhancing voxels were identified as belonging to the enhancing region of the lesion.
Results are shown in Fig. 2. This MS patient had 18 MS lesions out of which 3 of them had Gd enhancing regions. The dice score for (E+) and (L−) voxel prediction was 0.64 and 0.86 respectively. The top row in Fig. 2 shows a lesion which had only (L−) voxels and the proposed method successfully predicted that there were no (E+) voxels in the lesion. The second and third row of Fig. 2 shows performance of the method in presence of Gd enhanced voxels in the lesion. Our method identified the (E+) voxels in the lesion. However there were false positives around the Gd enhanced core of the lesions where non-enhancing voxels were identified as belonging to the enhancing region of the lesion.
4 Discussion
Our analysis shows that combining tissue microstructure information from multi-compartment T2 relaxometry and diffusion MRI model helps at yielding better prediction accuracy as compared to each feature being used alone. The higher TPR of features derived from MCDiff might be attributed to the fact that during an active BBB breakdown, there is a greater presence of inter and extra cellular fluid matters and inflammation as compared to non-enhancing parts of the lesion. The high-T2 water fraction from MCT2 model is capable of identifying higher inflammation. However, the medium-T2 water fraction, as described Sect. 2.1 is heterogeneous in terms of inter and extra-cellular fluids. However, MCDiff models are able to better explain such scenarios. The non-enhancing regions are demyelinated regions with inflammation which can be explained using MCT2 microstructure information.
Our study has certain limitations. The clinical data used in this work did not favour use of state of the art multi-compartment models. Single b-value data with 30 directions limited us to use a realtively simpler MCM model for MCDiff for reliable estimations. Higher number of echoes and shorter echo times for the T2 relaxometry data will facilitate MCT2 models. However, this study illustrates that even with clinical protocols, we have good detection of enhanced lesion regions using only MCT2 and MCDiff features.
5 Conclusion
We proposed a method to identify MS lesion voxels in which the brain tissues are undergoing active BBB breakdown using brain tissue microstructure information derived from advanced MRI techniques. The proposed method shows promise and motivation to work on improvement of the model from its current form. In the future work, we plan to have uncertainty measures on the predictions so that we can tackle the issue of false positive detection effectively. We also plan to test it on higher quality data to realize the true potential of the current framework.
References
Guttmann, C., Ahn, S.S., Hsu, L., Kikinis, R., Jolesz, F.A.: The evolution of multiple sclerosis lesions on serial MR. AJNR 16(7), 1481–1491 (1995)
Lassmann, H., Bruck, W., Lucchinetti, C.: Heterogeneity of multiple sclerosis pathogenesis: implications for diagnosis and therapy. Trends Mol. Med. 7(3), 115–121 (2001)
Brownlee, W.J., Hardy, T.A., Fazekas, F., Miller, D.H.: Diagnosis of multiple sclerosis: progress and challenges. Lancet 389(10076), 1336–1346 (2017)
Kanda, T., Ishii, K., Kawaguchi, H., Kitajima, K., Takenaka, D.: High signal intensity in the dentate nucleus and globus pallidus on unenhanced T1-weighted MR images: relationship with increasing cumulative dose of a gadolinium-based contrast material. Radiology 270(3), 834–841 (2013)
Olchowy, C., et al.: The presence of the gadolinium-based contrast agent depositions in the brain and symptoms of gadolinium neurotoxicity-a systematic review. PLoS One 12(2), e0171704 (2017)
Gulani, V., Calamante, F., Shellock, F.G., Kanal, E., Reeder, S.B., et al.: Gadolinium deposition in the brain: summary of evidence and recommendations. Lancet Neurol. 16(7), 564–570 (2017)
Panagiotaki, E., et al.: Compartment models of the diffusion MR signal in brain white matter: a taxonomy and comparison. Neuroimage 59(3), 2241–2254 (2012)
Laule, C., et al.: Magnetic resonance imaging of myelin. Neurotherapeutics 4(3), 460–484 (2007)
MacKay, A.L., Laule, C.: Magnetic resonance of myelin water: an in vivo marker for myelin. Brain Plast. 2(1), 71–91 (2016)
Lancaster, J.L., Andrews, T., Hardies, L.J., Dodd, S., Fox, P.T.: Three-pool model of white matter. J. Magn. Reson. Imaging 17(1), 1–10 (2003)
Crawley, A., Henkelman, R.: Errors in T2 estimation using multislice multiple-echo imaging. MRM 4(1), 34–47 (1987)
Prasloski, T., Mädler, B., Xiang, Q.S., MacKay, A., Jones, C.: Applications of stimulated echo correction to multicomponent T2 analysis. MRM 67(6), 1803–1814 (2012)
Layton, K.J., Morelande, M., Wright, D., Farrell, P.M., Moran, B., Johnston, L.A.: Modelling and estimation of multicomponent T2 distributions. IEEE TMI 32(8), 1423–1434 (2013)
Stamm, A., Commowick, O., Warfield, S.K., Vantini, S.: Comprehensive maximum likelihood estimation of diffusion compartment models towards reliable mapping of brain microstructure. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 622–630. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46726-9_72
Commowick, O., Wiest-Daesslé, N., Prima, S.: Block-matching strategies for rigid registration of multimodal medical images. In: 9th ISBI, pp. 700–703. IEEE (2012)
Ourselin, S., Roche, A., Prima, S., Ayache, N.: Block matching: a general framework to improve robustness of rigid registration of medical images. In: Delp, S.L., DiGoia, A.M., Jaramaz, B. (eds.) MICCAI 2000. LNCS, vol. 1935, pp. 557–566. Springer, Heidelberg (2000). https://doi.org/10.1007/978-3-540-40899-4_57
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20(3), 273–297 (1995)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Chatterjee, S., Commowick, O., Afacan, O., Warfield, S.K., Barillot, C. (2018). Identification of Gadolinium Contrast Enhanced Regions in MS Lesions Using Brain Tissue Microstructure Information Obtained from Diffusion and T2 Relaxometry MRI. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11072. Springer, Cham. https://doi.org/10.1007/978-3-030-00931-1_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-00931-1_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00930-4
Online ISBN: 978-3-030-00931-1
eBook Packages: Computer ScienceComputer Science (R0)